Pre-Training of Deep Bidirectional Protein Sequence Representations with Structural Information
Paper and Code
Nov 25, 2019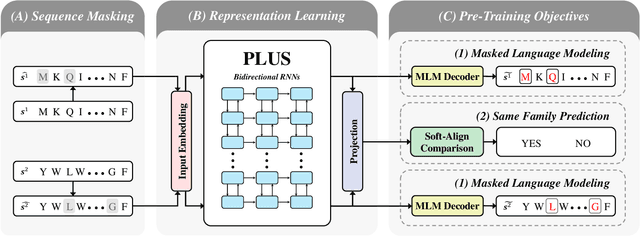
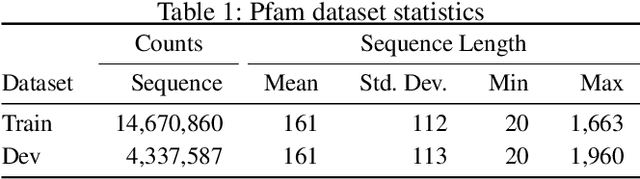
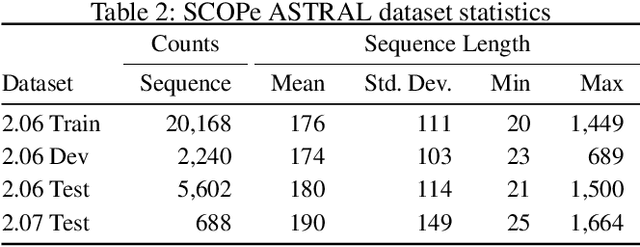
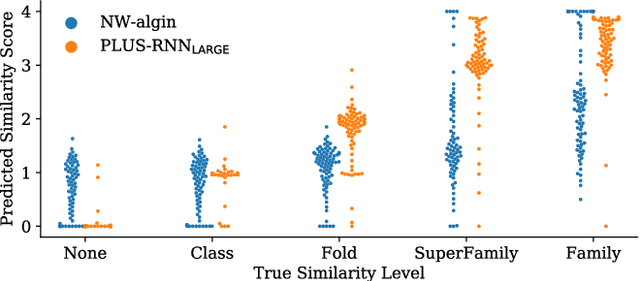
A structure of a protein has a direct impact on its properties and functions. However, identification of structural similarity directly from amino acid sequences remains as a challenging problem in computational biology. In this paper, we introduce a novel BERT-wise pre-training scheme for a protein sequence representation model called PLUS, which stands for Protein sequence representations Learned Using Structural information. As natural language representation models capture syntactic and semantic information of words from a large unlabeled text corpus, PLUS captures structural information of amino acids from a large weakly labeled protein database. Since the Transformer encoder, BERT's original model architecture, has a severe computational requirement to handle long sequences, we first propose to combine a bidirectional recurrent neural network with the BERT-wise pre-training scheme. PLUS is designed to learn protein representations with two pre-training objectives, i.e., masked language modeling and same family prediction. Then, the pre-trained model can be fine-tuned for a wide range of tasks without training randomly initialized task-specific models from scratch. It obtains new state-of-the-art results on both (1) protein-level and (2) amino-acid-level tasks, outperforming many task-specific algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge