Samuel J. Yang
Speech Intelligibility Classifiers from 550k Disordered Speech Samples
Mar 15, 2023Abstract:We developed dysarthric speech intelligibility classifiers on 551,176 disordered speech samples contributed by a diverse set of 468 speakers, with a range of self-reported speaking disorders and rated for their overall intelligibility on a five-point scale. We trained three models following different deep learning approaches and evaluated them on ~94K utterances from 100 speakers. We further found the models to generalize well (without further training) on the TORGO database (100% accuracy), UASpeech (0.93 correlation), ALS-TDI PMP (0.81 AUC) datasets as well as on a dataset of realistic unprompted speech we gathered (106 dysarthric and 76 control speakers,~2300 samples).
Physics-enhanced machine learning for virtual fluorescence microscopy
Apr 21, 2020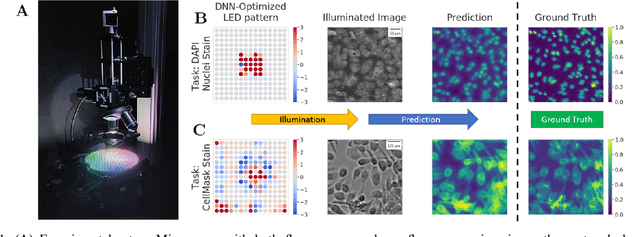
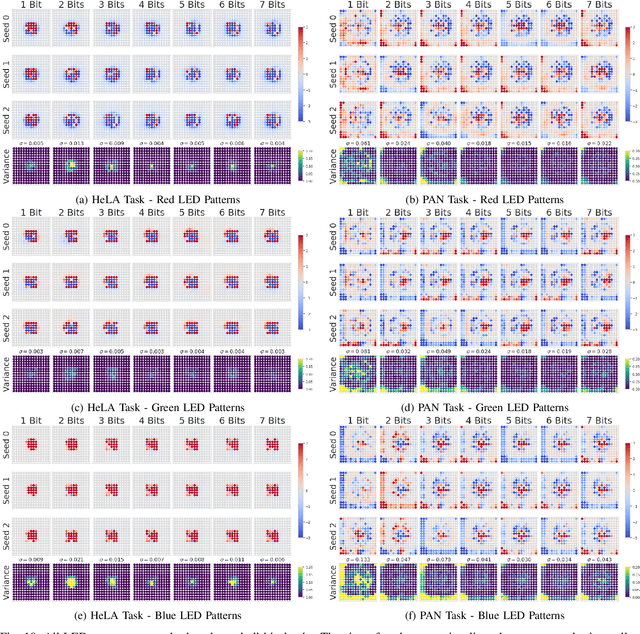
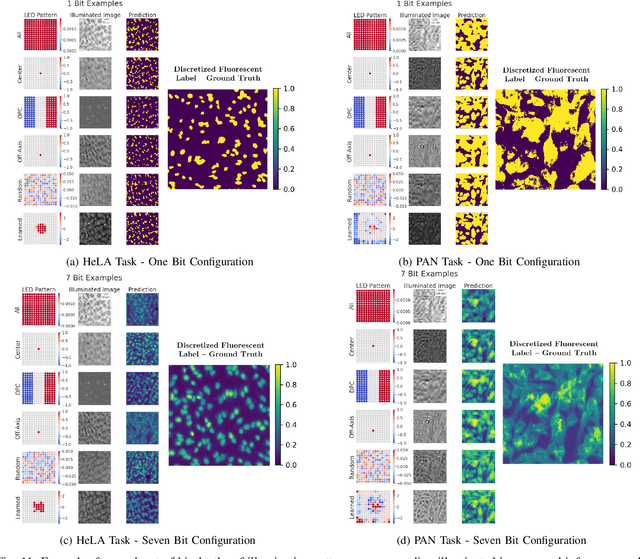
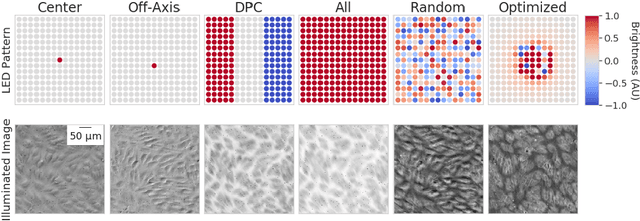
Abstract:This paper introduces a new method of data-driven microscope design for virtual fluorescence microscopy. Our results show that by including a model of illumination within the first layers of a deep convolutional neural network, it is possible to learn task-specific LED patterns that substantially improve the ability to infer fluorescence image information from unstained transmission microscopy images. We validated our method on two different experimental setups, with different magnifications and different sample types, to show a consistent improvement in performance as compared to conventional illumination methods. Additionally, to understand the importance of learned illumination on inference task, we varied the dynamic range of the fluorescent image targets (from one to seven bits), and showed that the margin of improvement for learned patterns increased with the information content of the target. This work demonstrates the power of programmable optical elements at enabling better machine learning algorithm performance and at providing physical insight into next generation of machine-controlled imaging systems.
Correcting Nuisance Variation using Wasserstein Distance
Nov 02, 2017
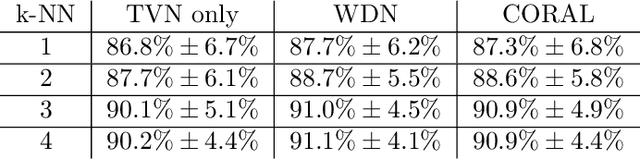
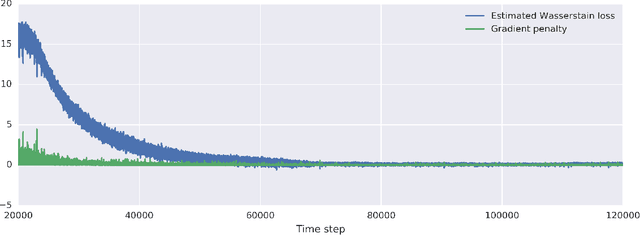
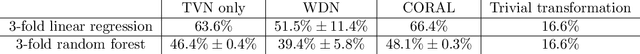
Abstract:Profiling cellular phenotypes from microscopic imaging can provide meaningful biological information resulting from various factors affecting the cells. One motivating application is drug development: morphological cell features can be captured from images, from which similarities between different drugs applied at different dosages can be quantified. The general approach is to find a function mapping the images to an embedding space of manageable dimensionality whose geometry captures relevant features of the input images. An important known issue for such methods is separating relevant biological signal from nuisance variation. For example, the embedding vectors tend to be more correlated for cells that were cultured and imaged during the same week than for cells from a different week, despite having identical drug compounds applied in both cases. In this case, the particular batch a set of experiments were conducted in constitutes the domain of the data; an ideal set of image embeddings should contain only the relevant biological information (e.g. drug effects). We develop a method for adjusting the image embeddings in order to `forget' domain-specific information while preserving relevant biological information. To do this, we minimize a loss function based on the Wasserstein distance. We find for our transformed embeddings (1) the underlying geometric structure is preserved and (2) less domain-specific information is present.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge