Qiyuan Hu
Efficient and generalizable prediction of molecular alterations in multiple cancer cohorts using H&E whole slide images
Jul 22, 2024Abstract:Molecular testing of tumor samples for targetable biomarkers is restricted by a lack of standardization, turnaround-time, cost, and tissue availability across cancer types. Additionally, targetable alterations of low prevalence may not be tested in routine workflows. Algorithms that predict DNA alterations from routinely generated hematoxylin and eosin (H&E)-stained images could prioritize samples for confirmatory molecular testing. Costs and the necessity of a large number of samples containing mutations limit approaches that train individual algorithms for each alteration. In this work, models were trained for simultaneous prediction of multiple DNA alterations from H&E images using a multi-task approach. Compared to biomarker-specific models, this approach performed better on average, with pronounced gains for rare mutations. The models reasonably generalized to independent temporal-holdout, externally-stained, and multi-site TCGA test sets. Additionally, whole slide image embeddings derived using multi-task models demonstrated strong performance in downstream tasks that were not a part of training. Overall, this is a promising approach to develop clinically useful algorithms that provide multiple actionable predictions from a single slide.
Development and Validation of a Deep Learning-Based Microsatellite Instability Predictor from Prostate Cancer Whole-Slide Images
Oct 12, 2023Abstract:Microsatellite instability-high (MSI-H) is a tumor agnostic biomarker for immune checkpoint inhibitor therapy. However, MSI status is not routinely tested in prostate cancer, in part due to low prevalence and assay cost. As such, prediction of MSI status from hematoxylin and eosin (H&E) stained whole-slide images (WSIs) could identify prostate cancer patients most likely to benefit from confirmatory testing and becoming eligible for immunotherapy. Prostate biopsies and surgical resections from de-identified records of consecutive prostate cancer patients referred to our institution were analyzed. Their MSI status was determined by next generation sequencing. Patients before a cutoff date were split into an algorithm development set (n=4015, MSI-H 1.8%) and a paired validation set (n=173, MSI-H 19.7%) that consisted of two serial sections from each sample, one stained and scanned internally and the other at an external site. Patients after the cutoff date formed the temporal validation set (n=1350, MSI-H 2.3%). Attention-based multiple instance learning models were trained to predict MSI-H from H&E WSIs. The MSI-H predictor achieved area under the receiver operating characteristic curve values of 0.78 (95% CI [0.69-0.86]), 0.72 (95% CI [0.63-0.81]), and 0.72 (95% CI [0.62-0.82]) on the internally prepared, externally prepared, and temporal validation sets, respectively. While MSI-H status is significantly correlated with Gleason score, the model remained predictive within each Gleason score subgroup. In summary, we developed and validated an AI-based MSI-H diagnostic model on a large real-world cohort of routine H&E slides, which effectively generalized to externally stained and scanned samples and a temporally independent validation cohort. This algorithm has the potential to direct prostate cancer patients toward immunotherapy and to identify MSI-H cases secondary to Lynch syndrome.
Make-A-Video: Text-to-Video Generation without Text-Video Data
Sep 29, 2022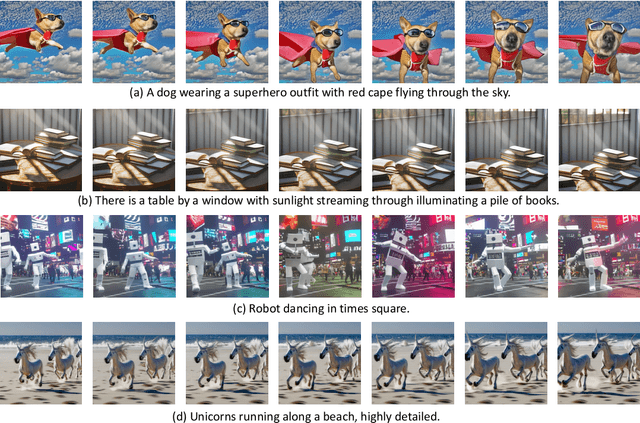

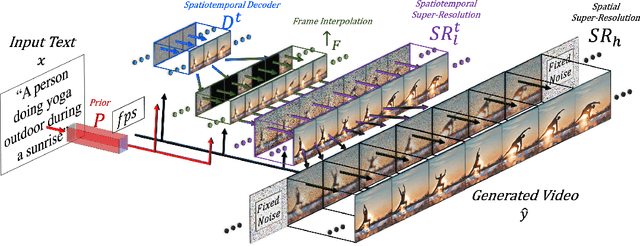
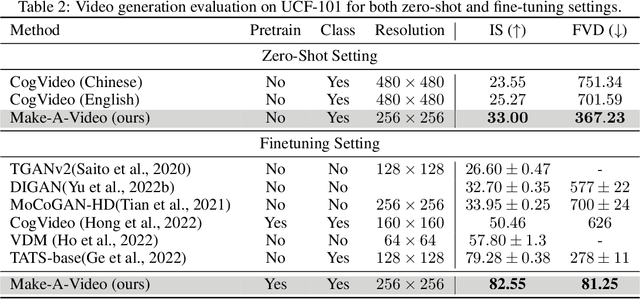
Abstract:We propose Make-A-Video -- an approach for directly translating the tremendous recent progress in Text-to-Image (T2I) generation to Text-to-Video (T2V). Our intuition is simple: learn what the world looks like and how it is described from paired text-image data, and learn how the world moves from unsupervised video footage. Make-A-Video has three advantages: (1) it accelerates training of the T2V model (it does not need to learn visual and multimodal representations from scratch), (2) it does not require paired text-video data, and (3) the generated videos inherit the vastness (diversity in aesthetic, fantastical depictions, etc.) of today's image generation models. We design a simple yet effective way to build on T2I models with novel and effective spatial-temporal modules. First, we decompose the full temporal U-Net and attention tensors and approximate them in space and time. Second, we design a spatial temporal pipeline to generate high resolution and frame rate videos with a video decoder, interpolation model and two super resolution models that can enable various applications besides T2V. In all aspects, spatial and temporal resolution, faithfulness to text, and quality, Make-A-Video sets the new state-of-the-art in text-to-video generation, as determined by both qualitative and quantitative measures.
MUGEN: A Playground for Video-Audio-Text Multimodal Understanding and GENeration
Apr 28, 2022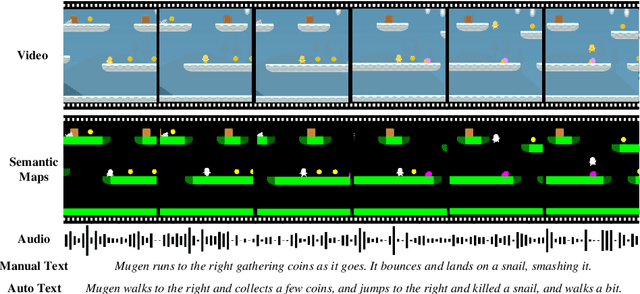
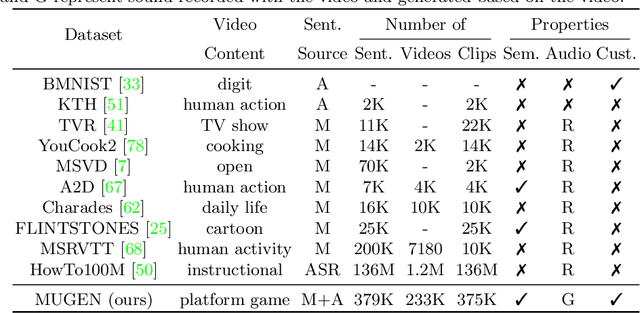
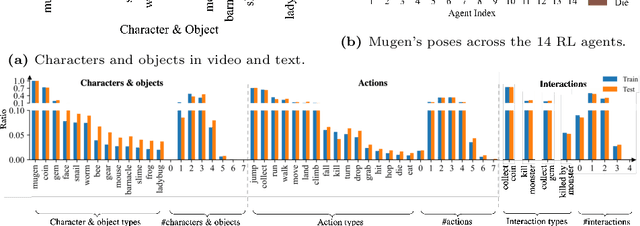
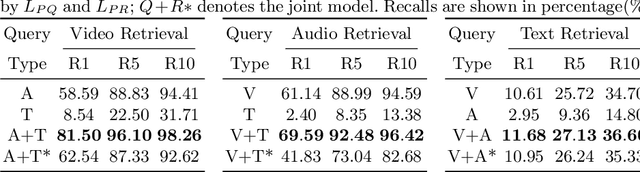
Abstract:Multimodal video-audio-text understanding and generation can benefit from datasets that are narrow but rich. The narrowness allows bite-sized challenges that the research community can make progress on. The richness ensures we are making progress along the core challenges. To this end, we present a large-scale video-audio-text dataset MUGEN, collected using the open-sourced platform game CoinRun [11]. We made substantial modifications to make the game richer by introducing audio and enabling new interactions. We trained RL agents with different objectives to navigate the game and interact with 13 objects and characters. This allows us to automatically extract a large collection of diverse videos and associated audio. We sample 375K video clips (3.2s each) and collect text descriptions from human annotators. Each video has additional annotations that are extracted automatically from the game engine, such as accurate semantic maps for each frame and templated textual descriptions. Altogether, MUGEN can help progress research in many tasks in multimodal understanding and generation. We benchmark representative approaches on tasks involving video-audio-text retrieval and generation. Our dataset and code are released at: https://mugen-org.github.io/.
Transfer Learning in 4D for Breast Cancer Diagnosis using Dynamic Contrast-Enhanced Magnetic Resonance Imaging
Nov 08, 2019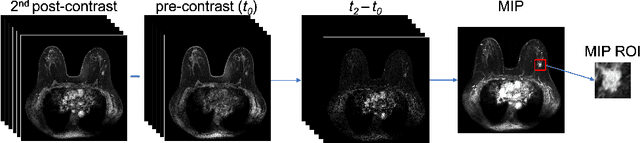
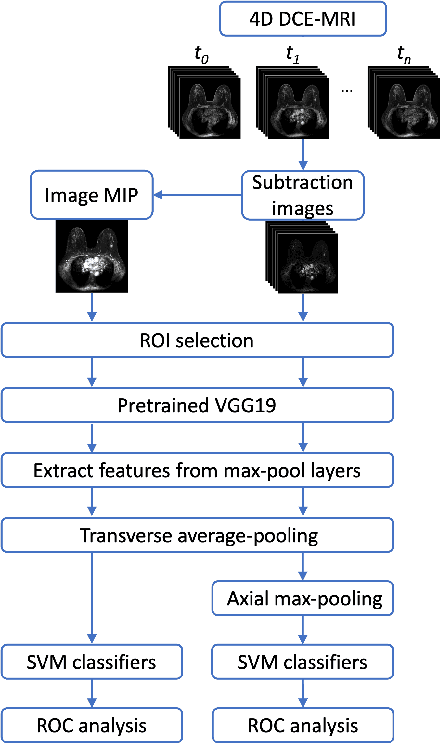
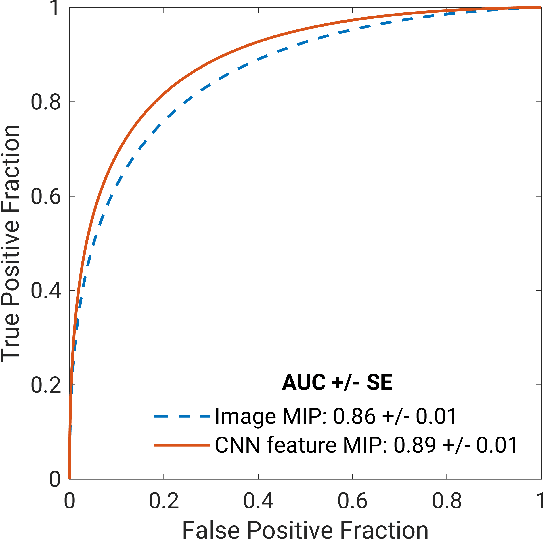
Abstract:Deep transfer learning using dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has shown strong predictive power in characterization of breast lesions. However, pretrained convolutional neural networks (CNNs) require 2D inputs, limiting the ability to exploit the rich 4D (volumetric and temporal) image information inherent in DCE-MRI that is clinically valuable for lesion assessment. Training 3D CNNs from scratch, a common method to utilize high-dimensional information in medical images, is computationally expensive and is not best suited for moderately sized healthcare datasets. Therefore, we propose a novel approach using transfer learning that incorporates the 4D information from DCE-MRI, where volumetric information is collapsed at feature level by max pooling along the projection perpendicular to the transverse slices and the temporal information is contained either in second-post contrast subtraction images. Our methodology yielded an area under the receiver operating characteristic curve of 0.89+/-0.01 on a dataset of 1161 breast lesions, significantly outperforming a previous approach that incorporates the 4D information in DCE-MRI by the use of maximum intensity projection (MIP) images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge