Heather M. Whitney
AI analysis of medical images at scale as a health disparities probe: a feasibility demonstration using chest radiographs
Apr 08, 2025



Abstract:Health disparities (differences in non-genetic conditions that influence health) can be associated with differences in burden of disease by groups within a population. Social determinants of health (SDOH) are domains such as health care access, dietary access, and economics frequently studied for potential association with health disparities. Evaluating SDOH-related phenotypes using routine medical images as data sources may enhance health disparities research. We developed a pipeline for using quantitative measures automatically extracted from medical images as inputs into health disparities index calculations. Our study focused on the use case of two SDOH demographic correlates (sex and race) and data extracted from chest radiographs of 1,571 unique patients. The likelihood of severe disease within the lung parenchyma from each image type, measured using an established deep learning model, was merged into a single numerical image-based phenotype for each patient. Patients were then separated into phenogroups by unsupervised clustering of the image-based phenotypes. The health rate for each phenogroup was defined as the median image-based phenotype for each SDOH used as inputs to four imaging-derived health disparities indices (iHDIs): one absolute measure (between-group variance) and three relative measures (index of disparity, Theil index, and mean log deviation). The iHDI measures demonstrated feasible values for each SDOH demographic correlate, showing potential for medical images to serve as a novel probe for health disparities. Large-scale AI analysis of medical images can serve as a probe for a novel data source for health disparities research.
Transfer Learning in 4D for Breast Cancer Diagnosis using Dynamic Contrast-Enhanced Magnetic Resonance Imaging
Nov 08, 2019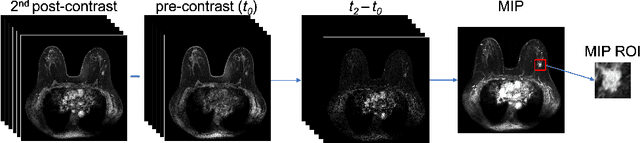
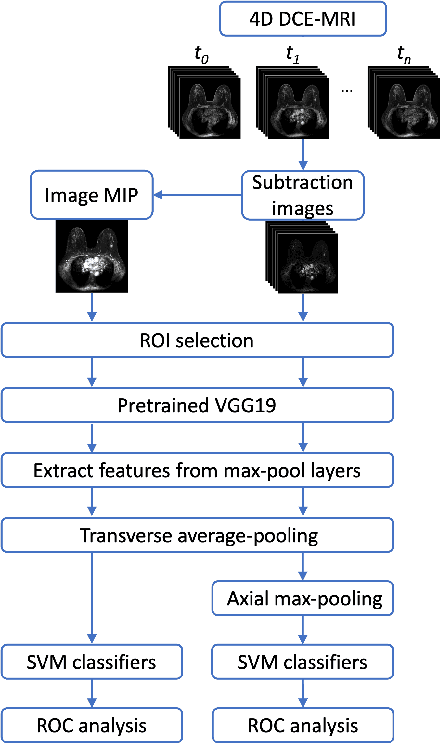
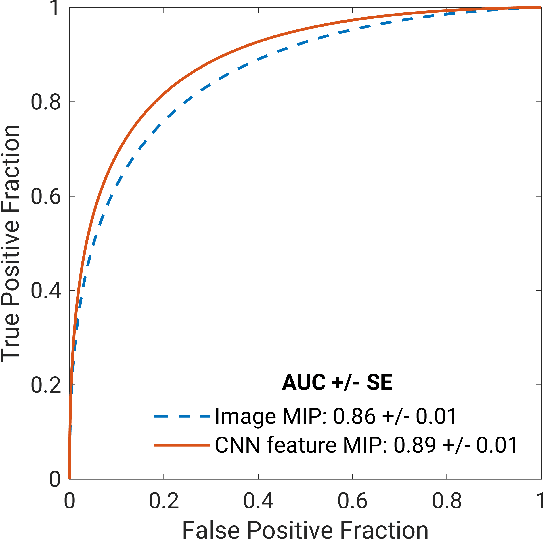
Abstract:Deep transfer learning using dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has shown strong predictive power in characterization of breast lesions. However, pretrained convolutional neural networks (CNNs) require 2D inputs, limiting the ability to exploit the rich 4D (volumetric and temporal) image information inherent in DCE-MRI that is clinically valuable for lesion assessment. Training 3D CNNs from scratch, a common method to utilize high-dimensional information in medical images, is computationally expensive and is not best suited for moderately sized healthcare datasets. Therefore, we propose a novel approach using transfer learning that incorporates the 4D information from DCE-MRI, where volumetric information is collapsed at feature level by max pooling along the projection perpendicular to the transverse slices and the temporal information is contained either in second-post contrast subtraction images. Our methodology yielded an area under the receiver operating characteristic curve of 0.89+/-0.01 on a dataset of 1161 breast lesions, significantly outperforming a previous approach that incorporates the 4D information in DCE-MRI by the use of maximum intensity projection (MIP) images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge