Aïcha BenTaieb
Development and Validation of a Deep Learning-Based Microsatellite Instability Predictor from Prostate Cancer Whole-Slide Images
Oct 12, 2023Abstract:Microsatellite instability-high (MSI-H) is a tumor agnostic biomarker for immune checkpoint inhibitor therapy. However, MSI status is not routinely tested in prostate cancer, in part due to low prevalence and assay cost. As such, prediction of MSI status from hematoxylin and eosin (H&E) stained whole-slide images (WSIs) could identify prostate cancer patients most likely to benefit from confirmatory testing and becoming eligible for immunotherapy. Prostate biopsies and surgical resections from de-identified records of consecutive prostate cancer patients referred to our institution were analyzed. Their MSI status was determined by next generation sequencing. Patients before a cutoff date were split into an algorithm development set (n=4015, MSI-H 1.8%) and a paired validation set (n=173, MSI-H 19.7%) that consisted of two serial sections from each sample, one stained and scanned internally and the other at an external site. Patients after the cutoff date formed the temporal validation set (n=1350, MSI-H 2.3%). Attention-based multiple instance learning models were trained to predict MSI-H from H&E WSIs. The MSI-H predictor achieved area under the receiver operating characteristic curve values of 0.78 (95% CI [0.69-0.86]), 0.72 (95% CI [0.63-0.81]), and 0.72 (95% CI [0.62-0.82]) on the internally prepared, externally prepared, and temporal validation sets, respectively. While MSI-H status is significantly correlated with Gleason score, the model remained predictive within each Gleason score subgroup. In summary, we developed and validated an AI-based MSI-H diagnostic model on a large real-world cohort of routine H&E slides, which effectively generalized to externally stained and scanned samples and a temporally independent validation cohort. This algorithm has the potential to direct prostate cancer patients toward immunotherapy and to identify MSI-H cases secondary to Lynch syndrome.
AI-augmented histopathologic review using image analysis to optimize DNA yield and tumor purity from FFPE slides
Apr 07, 2022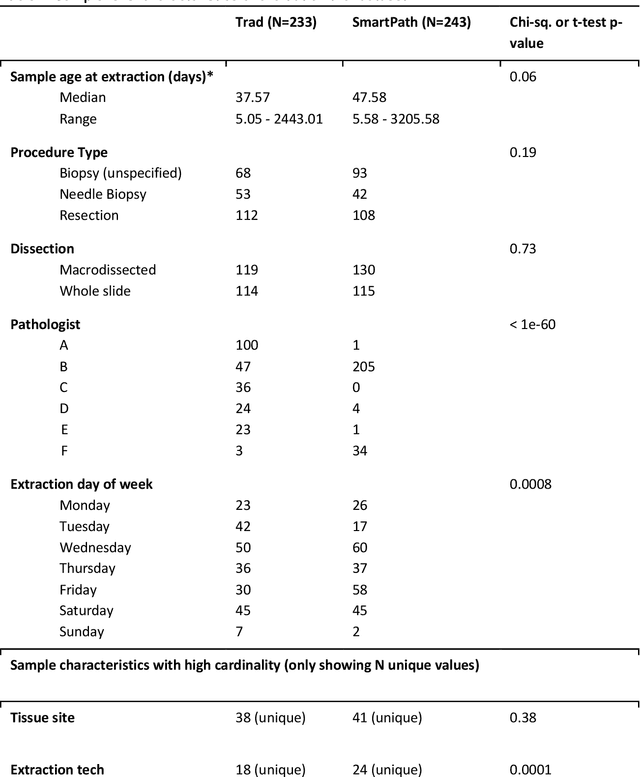

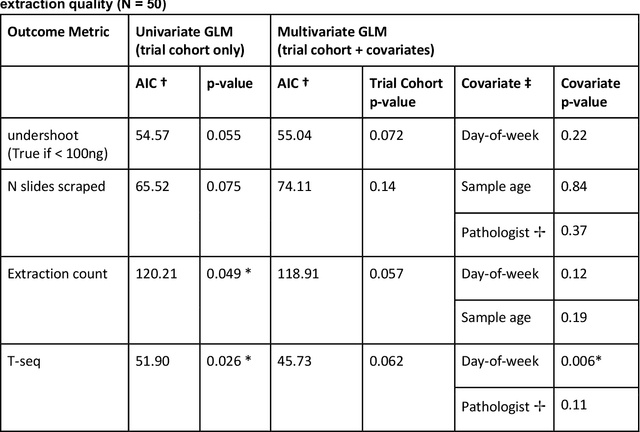

Abstract:To achieve minimum DNA input and tumor purity requirements for next-generation sequencing (NGS), pathologists visually estimate macrodissection and slide count decisions. Misestimation may cause tissue waste and increased laboratory costs. We developed an AI-augmented smart pathology review system (SmartPath) to empower pathologists with quantitative metrics for determining tissue extraction parameters. Using digitized H&E-stained FFPE slides as inputs, SmartPath segments tumors, extracts cell-based features, and suggests macrodissection areas. To predict DNA yield per slide, the extracted features are correlated with known DNA yields. Then, a pathologist-defined target yield divided by the predicted DNA yield/slide gives the number of slides to scrape. Following model development, an internal validation trial was conducted within the Tempus Labs molecular sequencing laboratory. We evaluated our system on 501 clinical colorectal cancer slides, where half received SmartPath-augmented review and half traditional pathologist review. The SmartPath cohort had 25% more DNA yields within a desired target range of 100-2000ng. The SmartPath system recommended fewer slides to scrape for large tissue sections, saving tissue in these cases. Conversely, SmartPath recommended more slides to scrape for samples with scant tissue sections, helping prevent costly re-extraction due to insufficient extraction yield. A statistical analysis was performed to measure the impact of covariates on the results, offering insights on how to improve future applications of SmartPath. Overall, the study demonstrated that AI-augmented histopathologic review using SmartPath could decrease tissue waste, sequencing time, and laboratory costs by optimizing DNA yields and tumor purity.
Deep Learning Models for Digital Pathology
Oct 29, 2019



Abstract:Histopathology images; microscopy images of stained tissue biopsies contain fundamental prognostic information that forms the foundation of pathological analysis and diagnostic medicine. However, diagnostics from histopathology images generally rely on a visual cognitive assessment of tissue slides which implies an inherent element of interpretation and hence subjectivity. Access to digitized histopathology images enabled the development of computational systems aiming at reducing manual intervention and automating parts of pathologists' workflow. Specifically, applications of deep learning to histopathology image analysis now offer opportunities for better quantitative modeling of disease appearance and hence possibly improved prediction of disease aggressiveness and patient outcome. However digitized histopathology tissue slides are unique in a variety of ways and come with their own set of computational challenges. In this survey, we summarize the different challenges facing computational systems for digital pathology and provide a review of state-of-the-art works that developed deep learning-based solutions for the predictive modeling of histopathology images from a detection, stain normalization, segmentation, and tissue classification perspective. We then discuss the challenges facing the validation and integration of such deep learning-based computational systems in clinical workflow and reflect on future opportunities for histopathology derived image measurements and better predictive modeling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge