Caleb Willis
AI-augmented histopathologic review using image analysis to optimize DNA yield and tumor purity from FFPE slides
Apr 07, 2022

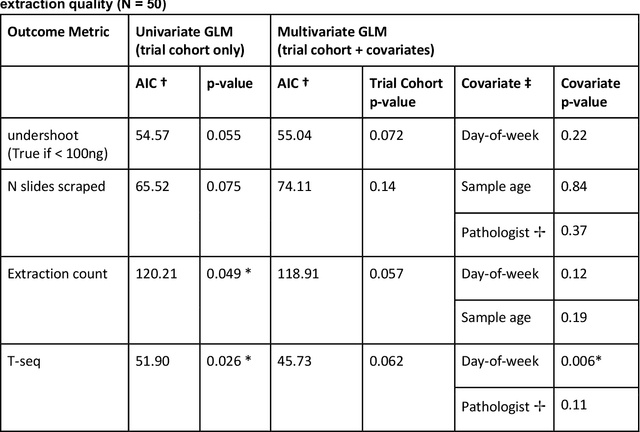

Abstract:To achieve minimum DNA input and tumor purity requirements for next-generation sequencing (NGS), pathologists visually estimate macrodissection and slide count decisions. Misestimation may cause tissue waste and increased laboratory costs. We developed an AI-augmented smart pathology review system (SmartPath) to empower pathologists with quantitative metrics for determining tissue extraction parameters. Using digitized H&E-stained FFPE slides as inputs, SmartPath segments tumors, extracts cell-based features, and suggests macrodissection areas. To predict DNA yield per slide, the extracted features are correlated with known DNA yields. Then, a pathologist-defined target yield divided by the predicted DNA yield/slide gives the number of slides to scrape. Following model development, an internal validation trial was conducted within the Tempus Labs molecular sequencing laboratory. We evaluated our system on 501 clinical colorectal cancer slides, where half received SmartPath-augmented review and half traditional pathologist review. The SmartPath cohort had 25% more DNA yields within a desired target range of 100-2000ng. The SmartPath system recommended fewer slides to scrape for large tissue sections, saving tissue in these cases. Conversely, SmartPath recommended more slides to scrape for samples with scant tissue sections, helping prevent costly re-extraction due to insufficient extraction yield. A statistical analysis was performed to measure the impact of covariates on the results, offering insights on how to improve future applications of SmartPath. Overall, the study demonstrated that AI-augmented histopathologic review using SmartPath could decrease tissue waste, sequencing time, and laboratory costs by optimizing DNA yields and tumor purity.
Deep Orthogonal Fusion: Multimodal Prognostic Biomarker Discovery Integrating Radiology, Pathology, Genomic, and Clinical Data
Jul 01, 2021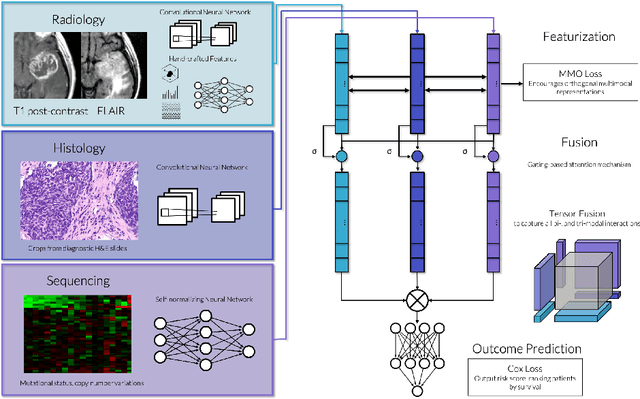
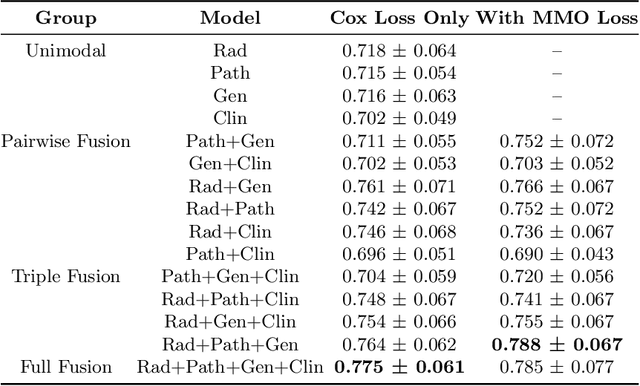
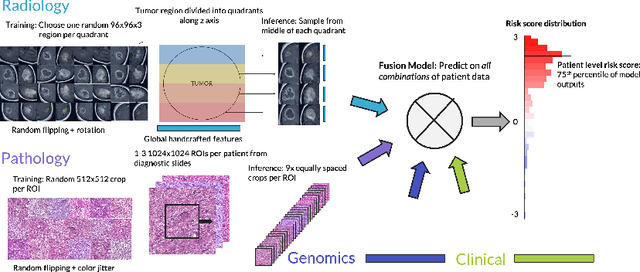
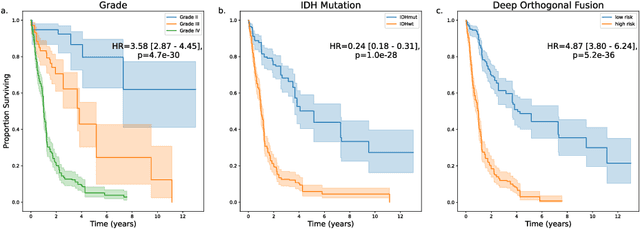
Abstract:Clinical decision-making in oncology involves multimodal data such as radiology scans, molecular profiling, histopathology slides, and clinical factors. Despite the importance of these modalities individually, no deep learning framework to date has combined them all to predict patient prognosis. Here, we predict the overall survival (OS) of glioma patients from diverse multimodal data with a Deep Orthogonal Fusion (DOF) model. The model learns to combine information from multiparametric MRI exams, biopsy-based modalities (such as H&E slide images and/or DNA sequencing), and clinical variables into a comprehensive multimodal risk score. Prognostic embeddings from each modality are learned and combined via attention-gated tensor fusion. To maximize the information gleaned from each modality, we introduce a multimodal orthogonalization (MMO) loss term that increases model performance by incentivizing constituent embeddings to be more complementary. DOF predicts OS in glioma patients with a median C-index of 0.788 +/- 0.067, significantly outperforming (p=0.023) the best performing unimodal model with a median C-index of 0.718 +/- 0.064. The prognostic model significantly stratifies glioma patients by OS within clinical subsets, adding further granularity to prognostic clinical grading and molecular subtyping.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge