Nikhil Shenoy
Implicit Delta Learning of High Fidelity Neural Network Potentials
Dec 08, 2024Abstract:Neural network potentials (NNPs) offer a fast and accurate alternative to ab-initio methods for molecular dynamics (MD) simulations but are hindered by the high cost of training data from high-fidelity Quantum Mechanics (QM) methods. Our work introduces the Implicit Delta Learning (IDLe) method, which reduces the need for high-fidelity QM data by leveraging cheaper semi-empirical QM computations without compromising NNP accuracy or inference cost. IDLe employs an end-to-end multi-task architecture with fidelity-specific heads that decode energies based on a shared latent representation of the input atomistic system. In various settings, IDLe achieves the same accuracy as single high-fidelity baselines while using up to 50x less high-fidelity data. This result could significantly reduce data generation cost and consequently enhance accuracy and generalization, and expand chemical coverage for NNPs, advancing MD simulations for material science and drug discovery. Additionally, we provide a novel set of 11 million semi-empirical QM calculations to support future multi-fidelity NNP modeling.
OpenQDC: Open Quantum Data Commons
Nov 29, 2024



Abstract:Machine Learning Interatomic Potentials (MLIPs) are a highly promising alternative to force-fields for molecular dynamics (MD) simulations, offering precise and rapid energy and force calculations. However, Quantum-Mechanical (QM) datasets, crucial for MLIPs, are fragmented across various repositories, hindering accessibility and model development. We introduce the openQDC package, consolidating 37 QM datasets from over 250 quantum methods and 400 million geometries into a single, accessible resource. These datasets are meticulously preprocessed, and standardized for MLIP training, covering a wide range of chemical elements and interactions relevant in organic chemistry. OpenQDC includes tools for normalization and integration, easily accessible via Python. Experiments with well-known architectures like SchNet, TorchMD-Net, and DimeNet reveal challenges for those architectures and constitute a leaderboard to accelerate benchmarking and guide novel algorithms development. Continuously adding datasets to OpenQDC will democratize QM dataset access, foster more collaboration and innovation, enhance MLIP development, and support their adoption in the MD field.
ET-Flow: Equivariant Flow-Matching for Molecular Conformer Generation
Oct 29, 2024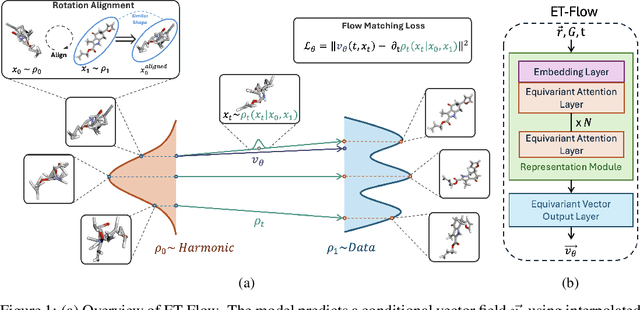
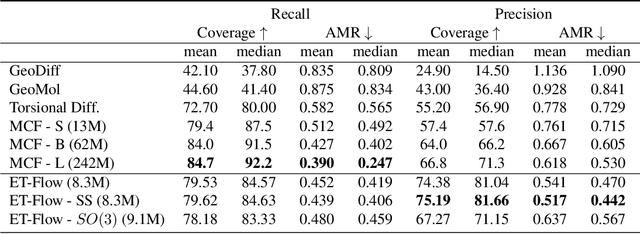
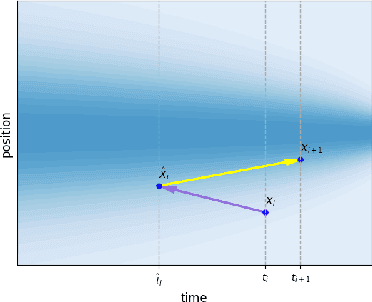
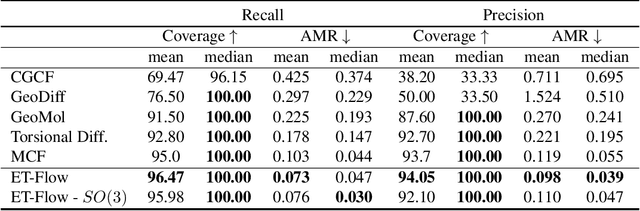
Abstract:Predicting low-energy molecular conformations given a molecular graph is an important but challenging task in computational drug discovery. Existing state-of-the-art approaches either resort to large scale transformer-based models that diffuse over conformer fields, or use computationally expensive methods to generate initial structures and diffuse over torsion angles. In this work, we introduce Equivariant Transformer Flow (ET-Flow). We showcase that a well-designed flow matching approach with equivariance and harmonic prior alleviates the need for complex internal geometry calculations and large architectures, contrary to the prevailing methods in the field. Our approach results in a straightforward and scalable method that directly operates on all-atom coordinates with minimal assumptions. With the advantages of equivariance and flow matching, ET-Flow significantly increases the precision and physical validity of the generated conformers, while being a lighter model and faster at inference. Code is available https://github.com/shenoynikhil/ETFlow.
Role of Structural and Conformational Diversity for Machine Learning Potentials
Oct 30, 2023



Abstract:In the field of Machine Learning Interatomic Potentials (MLIPs), understanding the intricate relationship between data biases, specifically conformational and structural diversity, and model generalization is critical in improving the quality of Quantum Mechanics (QM) data generation efforts. We investigate these dynamics through two distinct experiments: a fixed budget one, where the dataset size remains constant, and a fixed molecular set one, which focuses on fixed structural diversity while varying conformational diversity. Our results reveal nuanced patterns in generalization metrics. Notably, for optimal structural and conformational generalization, a careful balance between structural and conformational diversity is required, but existing QM datasets do not meet that trade-off. Additionally, our results highlight the limitation of the MLIP models at generalizing beyond their training distribution, emphasizing the importance of defining applicability domain during model deployment. These findings provide valuable insights and guidelines for QM data generation efforts.
A Case for Rejection in Low Resource ML Deployment
Aug 15, 2022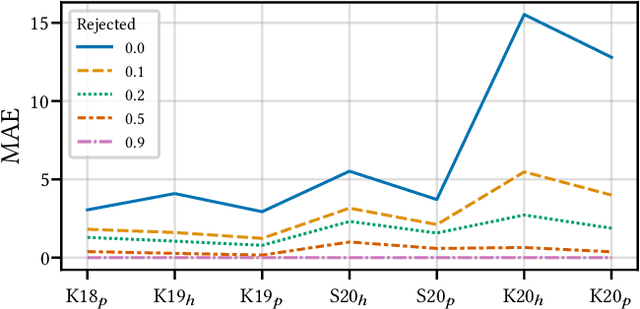
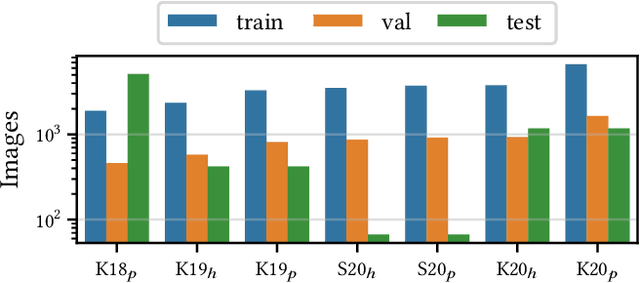
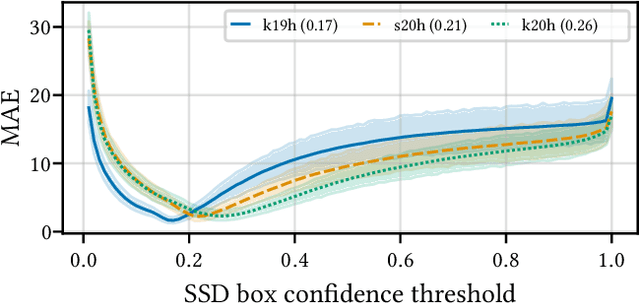
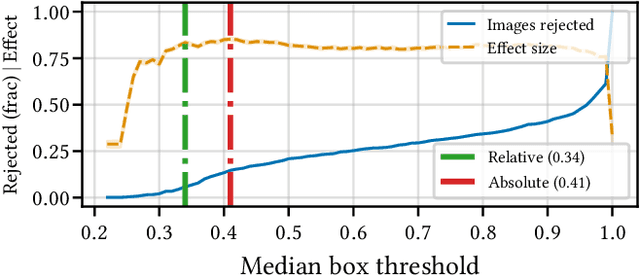
Abstract:Building reliable AI decision support systems requires a robust set of data on which to train models; both with respect to quantity and diversity. Obtaining such datasets can be difficult in resource limited settings, or for applications in early stages of deployment. Sample rejection is one way to work around this challenge, however much of the existing work in this area is ill-suited for such scenarios. This paper substantiates that position and proposes a simple solution as a proof of concept baseline.
Impact of data-splits on generalization: Identifying COVID-19 from cough and context
Jun 05, 2021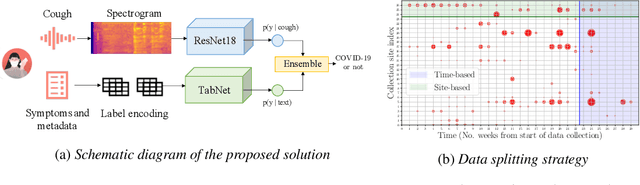
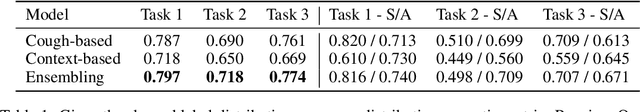
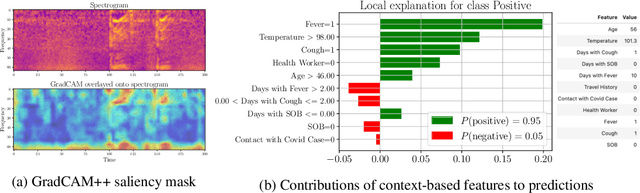
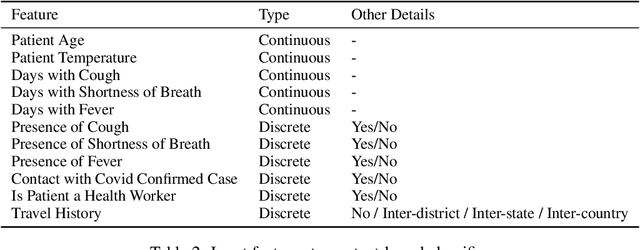
Abstract:Rapidly scaling screening, testing and quarantine has shown to be an effective strategy to combat the COVID-19 pandemic. We consider the application of deep learning techniques to distinguish individuals with COVID from non-COVID by using data acquirable from a phone. Using cough and context (symptoms and meta-data) represent such a promising approach. Several independent works in this direction have shown promising results. However, none of them report performance across clinically relevant data splits. Specifically, the performance where the development and test sets are split in time (retrospective validation) and across sites (broad validation). Although there is meaningful generalization across these splits the performance significantly varies (up to 0.1 AUC score). In addition, we study the performance of symptomatic and asymptomatic individuals across these three splits. Finally, we show that our model focuses on meaningful features of the input, cough bouts for cough and relevant symptoms for context. The code and checkpoints are available at https://github.com/WadhwaniAI/cough-against-covid
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge