Mingya Zhang
WT-BCP: Wavelet Transform based Bidirectional Copy-Paste for Semi-Supervised Medical Image Segmentation
Apr 20, 2025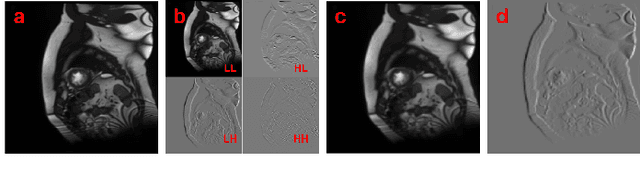
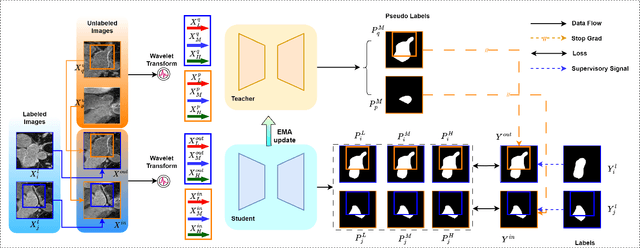
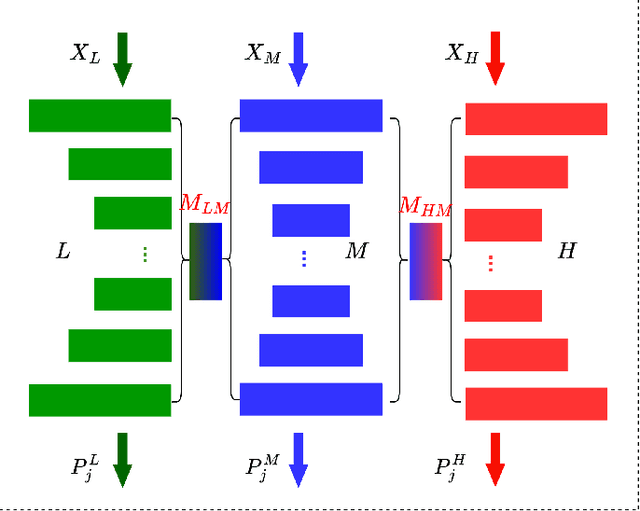
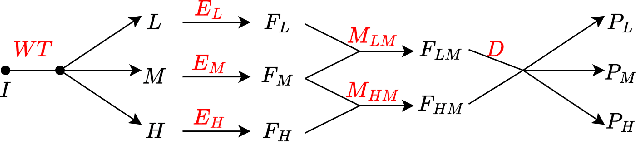
Abstract:Semi-supervised medical image segmentation (SSMIS) shows promise in reducing reliance on scarce labeled medical data. However, SSMIS field confronts challenges such as distribution mismatches between labeled and unlabeled data, artificial perturbations causing training biases, and inadequate use of raw image information, especially low-frequency (LF) and high-frequency (HF) components.To address these challenges, we propose a Wavelet Transform based Bidirectional Copy-Paste SSMIS framework, named WT-BCP, which improves upon the Mean Teacher approach. Our method enhances unlabeled data understanding by copying random crops between labeled and unlabeled images and employs WT to extract LF and HF details.We propose a multi-input and multi-output model named XNet-Plus, to receive the fused information after WT. Moreover, consistency training among multiple outputs helps to mitigate learning biases introduced by artificial perturbations. During consistency training, the mixed images resulting from WT are fed into both models, with the student model's output being supervised by pseudo-labels and ground-truth. Extensive experiments conducted on 2D and 3D datasets confirm the effectiveness of our model.Code: https://github.com/simzhangbest/WT-BCP.
Fraesormer: Learning Adaptive Sparse Transformer for Efficient Food Recognition
Mar 15, 2025Abstract:In recent years, Transformer has witnessed significant progress in food recognition. However, most existing approaches still face two critical challenges in lightweight food recognition: (1) the quadratic complexity and redundant feature representation from interactions with irrelevant tokens; (2) static feature recognition and single-scale representation, which overlook the unstructured, non-fixed nature of food images and the need for multi-scale features. To address these, we propose an adaptive and efficient sparse Transformer architecture (Fraesormer) with two core designs: Adaptive Top-k Sparse Partial Attention (ATK-SPA) and Hierarchical Scale-Sensitive Feature Gating Network (HSSFGN). ATK-SPA uses a learnable Gated Dynamic Top-K Operator (GDTKO) to retain critical attention scores, filtering low query-key matches that hinder feature aggregation. It also introduces a partial channel mechanism to reduce redundancy and promote expert information flow, enabling local-global collaborative modeling. HSSFGN employs gating mechanism to achieve multi-scale feature representation, enhancing contextual semantic information. Extensive experiments show that Fraesormer outperforms state-of-the-art methods. code is available at https://zs1314.github.io/Fraesormer.
Learning Dual-Domain Multi-Scale Representations for Single Image Deraining
Mar 15, 2025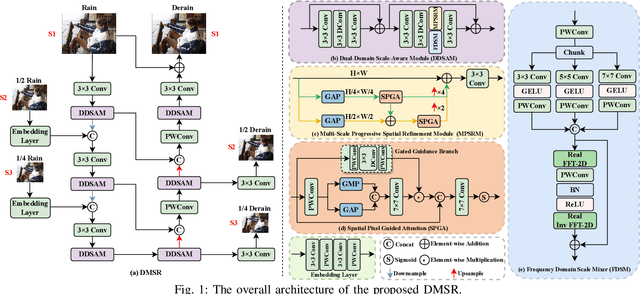
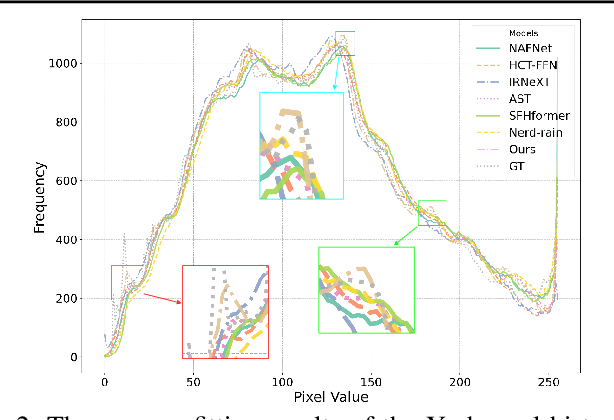
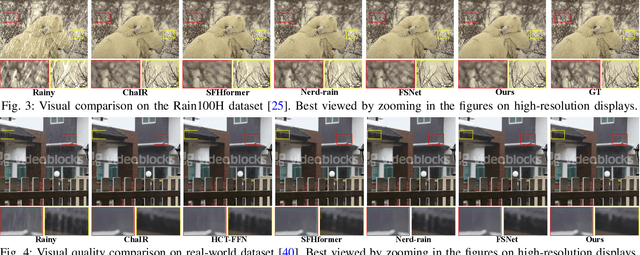
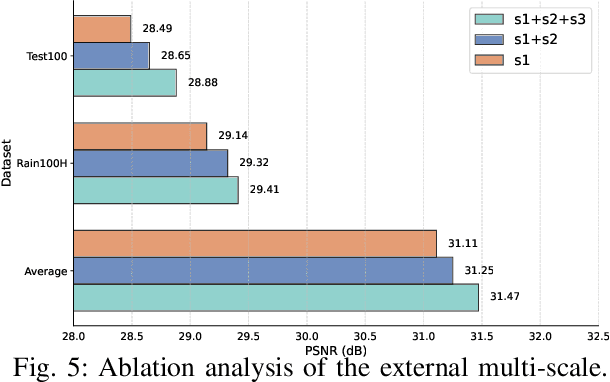
Abstract:Existing image deraining methods typically rely on single-input, single-output, and single-scale architectures, which overlook the joint multi-scale information between external and internal features. Furthermore, single-domain representations are often too restrictive, limiting their ability to handle the complexities of real-world rain scenarios. To address these challenges, we propose a novel Dual-Domain Multi-Scale Representation Network (DMSR). The key idea is to exploit joint multi-scale representations from both external and internal domains in parallel while leveraging the strengths of both spatial and frequency domains to capture more comprehensive properties. Specifically, our method consists of two main components: the Multi-Scale Progressive Spatial Refinement Module (MPSRM) and the Frequency Domain Scale Mixer (FDSM). The MPSRM enables the interaction and coupling of multi-scale expert information within the internal domain using a hierarchical modulation and fusion strategy. The FDSM extracts multi-scale local information in the spatial domain, while also modeling global dependencies in the frequency domain. Extensive experiments show that our model achieves state-of-the-art performance across six benchmark datasets.
Spectral Enhancement and Pseudo-Anchor Guidance for Infrared-Visible Person Re-Identification
Dec 26, 2024



Abstract:The development of deep learning has facilitated the application of person re-identification (ReID) technology in intelligent security. Visible-infrared person re-identification (VI-ReID) aims to match pedestrians across infrared and visible modality images enabling 24-hour surveillance. Current studies relying on unsupervised modality transformations as well as inefficient embedding constraints to bridge the spectral differences between infrared and visible images, however, limit their potential performance. To tackle the limitations of the above approaches, this paper introduces a simple yet effective Spectral Enhancement and Pseudo-anchor Guidance Network, named SEPG-Net. Specifically, we propose a more homogeneous spectral enhancement scheme based on frequency domain information and greyscale space, which avoids the information loss typically caused by inefficient modality transformations. Further, a Pseudo Anchor-guided Bidirectional Aggregation (PABA) loss is introduced to bridge local modality discrepancies while better preserving discriminative identity embeddings. Experimental results on two public benchmark datasets demonstrate the superior performance of SEPG-Net against other state-of-the-art methods. The code is available at https://github.com/1024AILab/ReID-SEPG.
SkinMamba: A Precision Skin Lesion Segmentation Architecture with Cross-Scale Global State Modeling and Frequency Boundary Guidance
Sep 17, 2024



Abstract:Skin lesion segmentation is a crucial method for identifying early skin cancer. In recent years, both convolutional neural network (CNN) and Transformer-based methods have been widely applied. Moreover, combining CNN and Transformer effectively integrates global and local relationships, but remains limited by the quadratic complexity of Transformer. To address this, we propose a hybrid architecture based on Mamba and CNN, called SkinMamba. It maintains linear complexity while offering powerful long-range dependency modeling and local feature extraction capabilities. Specifically, we introduce the Scale Residual State Space Block (SRSSB), which captures global contextual relationships and cross-scale information exchange at a macro level, enabling expert communication in a global state. This effectively addresses challenges in skin lesion segmentation related to varying lesion sizes and inconspicuous target areas. Additionally, to mitigate boundary blurring and information loss during model downsampling, we introduce the Frequency Boundary Guided Module (FBGM), providing sufficient boundary priors to guide precise boundary segmentation, while also using the retained information to assist the decoder in the decoding process. Finally, we conducted comparative and ablation experiments on two public lesion segmentation datasets (ISIC2017 and ISIC2018), and the results demonstrate the strong competitiveness of SkinMamba in skin lesion segmentation tasks. The code is available at https://github.com/zs1314/SkinMamba.
HMT-UNet: A hybird Mamba-Transformer Vision UNet for Medical Image Segmentation
Aug 21, 2024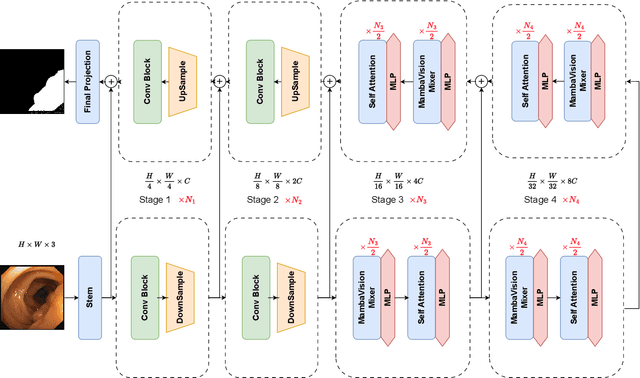
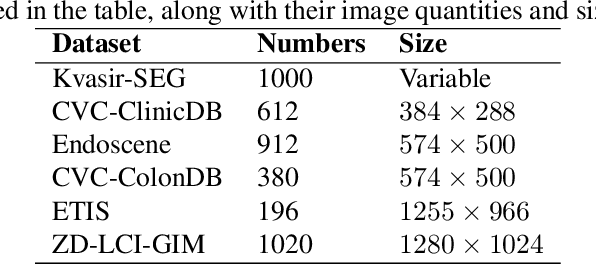
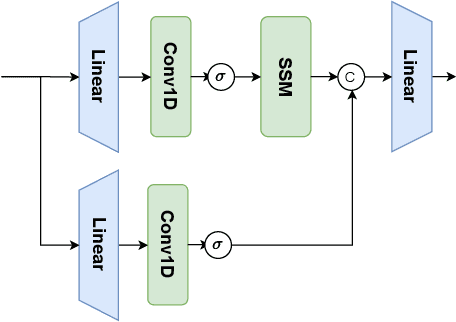
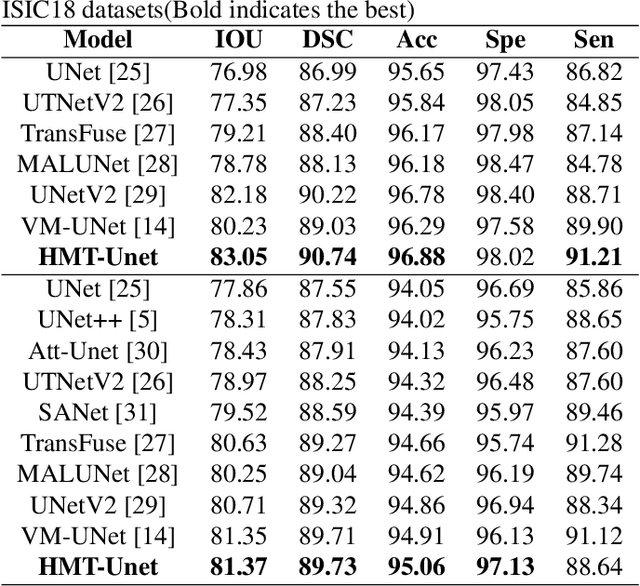
Abstract:In the field of medical image segmentation, models based on both CNN and Transformer have been thoroughly investigated. However, CNNs have limited modeling capabilities for long-range dependencies, making it challenging to exploit the semantic information within images fully. On the other hand, the quadratic computational complexity poses a challenge for Transformers. State Space Models (SSMs), such as Mamba, have been recognized as a promising method. They not only demonstrate superior performance in modeling long-range interactions, but also preserve a linear computational complexity. The hybrid mechanism of SSM (State Space Model) and Transformer, after meticulous design, can enhance its capability for efficient modeling of visual features. Extensive experiments have demonstrated that integrating the self-attention mechanism into the hybrid part behind the layers of Mamba's architecture can greatly improve the modeling capacity to capture long-range spatial dependencies. In this paper, leveraging the hybrid mechanism of SSM, we propose a U-shape architecture model for medical image segmentation, named Hybird Transformer vision Mamba UNet (HTM-UNet). We conduct comprehensive experiments on the ISIC17, ISIC18, CVC-300, CVC-ClinicDB, Kvasir, CVC-ColonDB, ETIS-Larib PolypDB public datasets and ZD-LCI-GIM private dataset. The results indicate that HTM-UNet exhibits competitive performance in medical image segmentation tasks. Our code is available at https://github.com/simzhangbest/HMT-Unet.
SAM2-PATH: A better segment anything model for semantic segmentation in digital pathology
Aug 07, 2024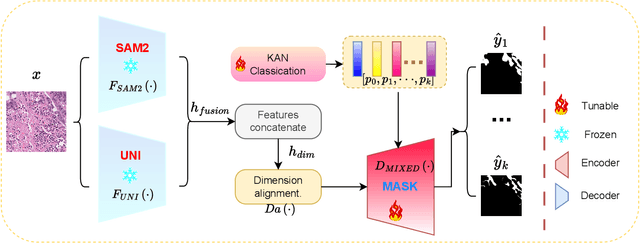
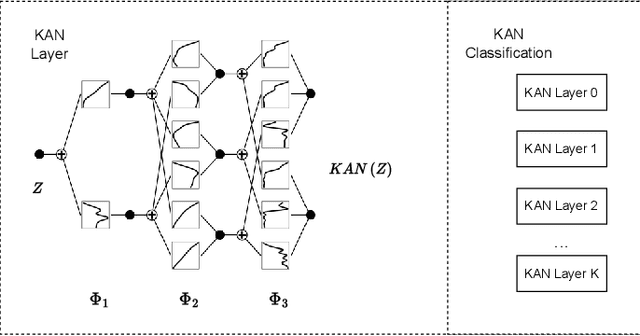
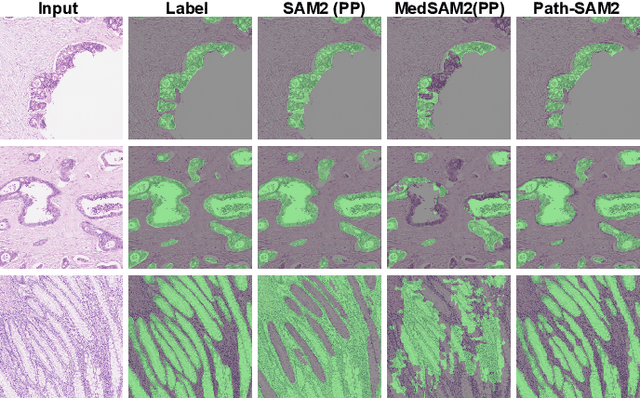
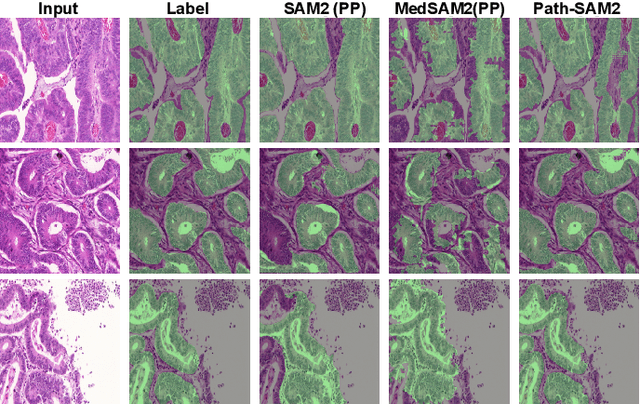
Abstract:The semantic segmentation task in pathology plays an indispensable role in assisting physicians in determining the condition of tissue lesions. Foundation models, such as the SAM (Segment Anything Model) and SAM2, exhibit exceptional performance in instance segmentation within everyday natural scenes. SAM-PATH has also achieved impressive results in semantic segmentation within the field of pathology. However, in computational pathology, the models mentioned above still have the following limitations. The pre-trained encoder models suffer from a scarcity of pathology image data; SAM and SAM2 are not suitable for semantic segmentation. In this paper, we have designed a trainable Kolmogorov-Arnold Networks(KAN) classification module within the SAM2 workflow, and we have introduced the largest pretrained vision encoder for histopathology (UNI) to date. Our proposed framework, SAM2-PATH, augments SAM2's capability to perform semantic segmentation in digital pathology autonomously, eliminating the need for human provided input prompts. The experimental results demonstrate that, after fine-tuning the KAN classification module and decoder, Our dataset has achieved competitive results on publicly available pathology data. The code has been open-sourced and can be found at the following address: https://github.com/simzhangbest/SAM2PATH.
VM-UNET-V2 Rethinking Vision Mamba UNet for Medical Image Segmentation
Mar 14, 2024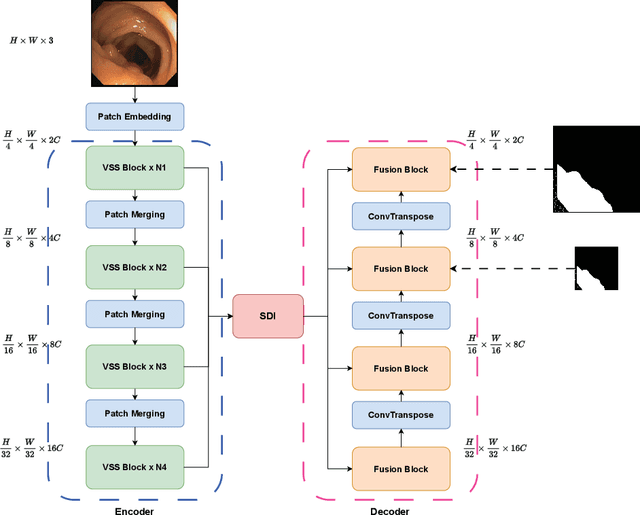
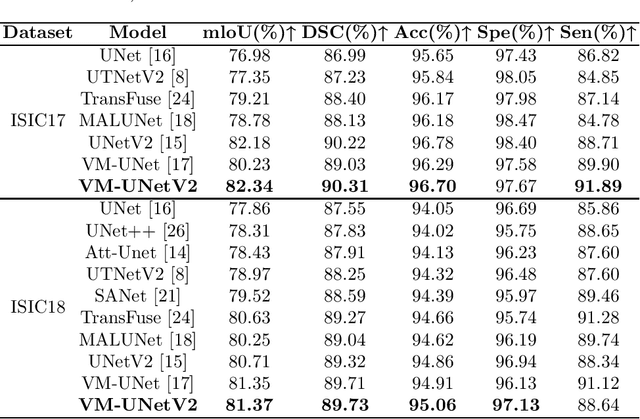
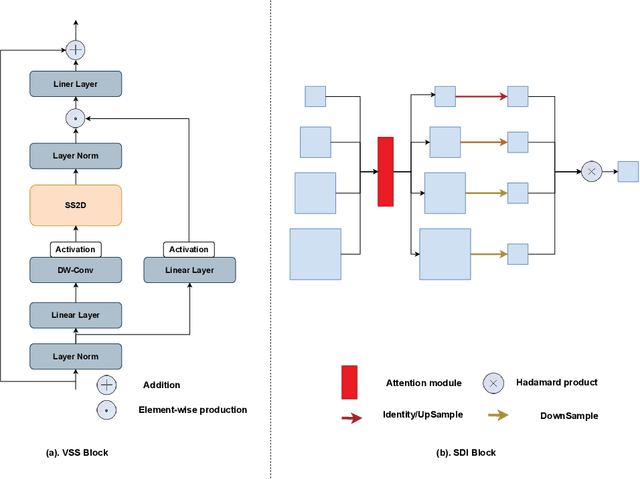
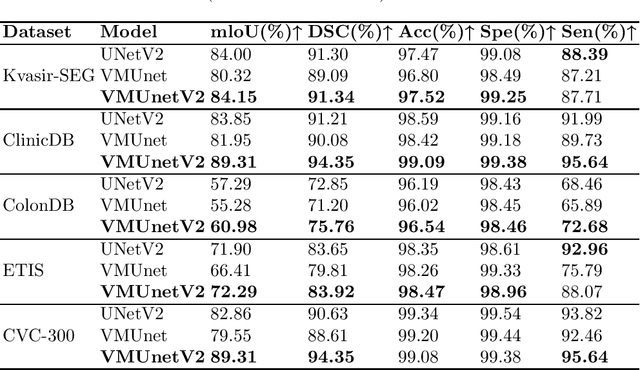
Abstract:In the field of medical image segmentation, models based on both CNN and Transformer have been thoroughly investigated. However, CNNs have limited modeling capabilities for long-range dependencies, making it challenging to exploit the semantic information within images fully. On the other hand, the quadratic computational complexity poses a challenge for Transformers. Recently, State Space Models (SSMs), such as Mamba, have been recognized as a promising method. They not only demonstrate superior performance in modeling long-range interactions, but also preserve a linear computational complexity. Inspired by the Mamba architecture, We proposed Vison Mamba-UNetV2, the Visual State Space (VSS) Block is introduced to capture extensive contextual information, the Semantics and Detail Infusion (SDI) is introduced to augment the infusion of low-level and high-level features. We conduct comprehensive experiments on the ISIC17, ISIC18, CVC-300, CVC-ClinicDB, Kvasir, CVC-ColonDB and ETIS-LaribPolypDB public datasets. The results indicate that VM-UNetV2 exhibits competitive performance in medical image segmentation tasks. Our code is available at https://github.com/nobodyplayer1/VM-UNetV2.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge