Shun Zou
Cross Paradigm Representation and Alignment Transformer for Image Deraining
Apr 23, 2025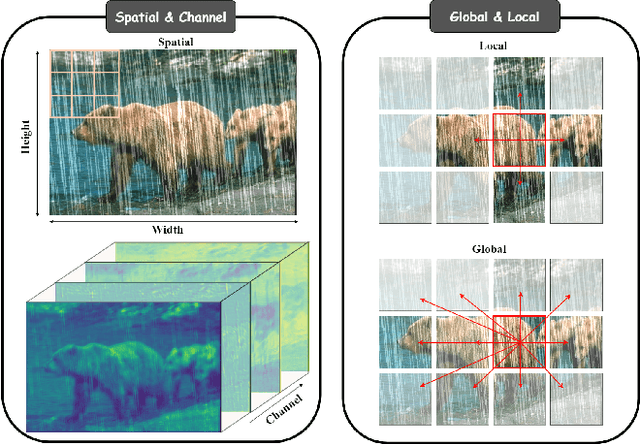
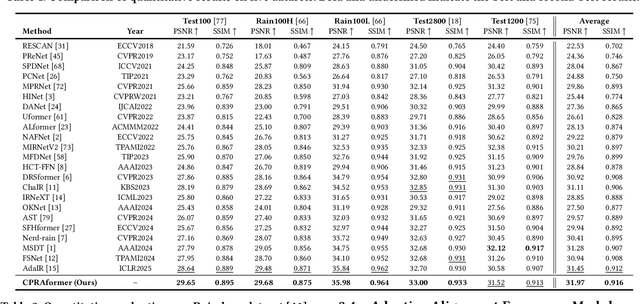
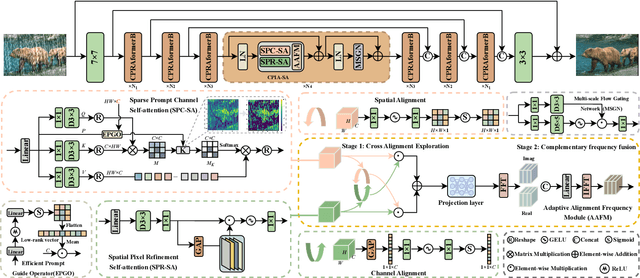
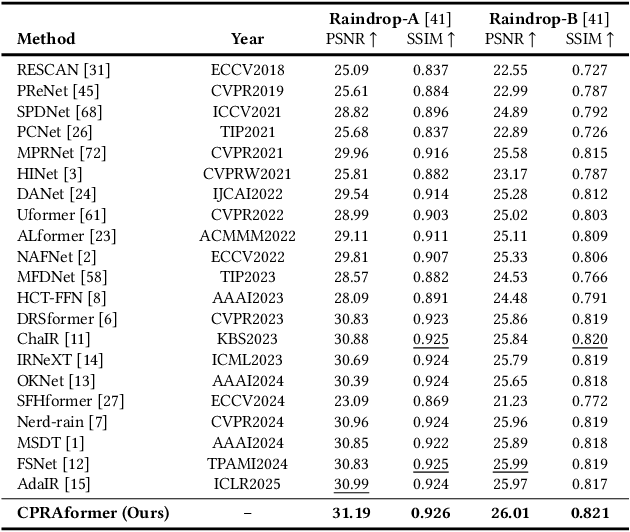
Abstract:Transformer-based networks have achieved strong performance in low-level vision tasks like image deraining by utilizing spatial or channel-wise self-attention. However, irregular rain patterns and complex geometric overlaps challenge single-paradigm architectures, necessitating a unified framework to integrate complementary global-local and spatial-channel representations. To address this, we propose a novel Cross Paradigm Representation and Alignment Transformer (CPRAformer). Its core idea is the hierarchical representation and alignment, leveraging the strengths of both paradigms (spatial-channel and global-local) to aid image reconstruction. It bridges the gap within and between paradigms, aligning and coordinating them to enable deep interaction and fusion of features. Specifically, we use two types of self-attention in the Transformer blocks: sparse prompt channel self-attention (SPC-SA) and spatial pixel refinement self-attention (SPR-SA). SPC-SA enhances global channel dependencies through dynamic sparsity, while SPR-SA focuses on spatial rain distribution and fine-grained texture recovery. To address the feature misalignment and knowledge differences between them, we introduce the Adaptive Alignment Frequency Module (AAFM), which aligns and interacts with features in a two-stage progressive manner, enabling adaptive guidance and complementarity. This reduces the information gap within and between paradigms. Through this unified cross-paradigm dynamic interaction framework, we achieve the extraction of the most valuable interactive fusion information from the two paradigms. Extensive experiments demonstrate that our model achieves state-of-the-art performance on eight benchmark datasets and further validates CPRAformer's robustness in other image restoration tasks and downstream applications.
Learning Dual-Domain Multi-Scale Representations for Single Image Deraining
Mar 15, 2025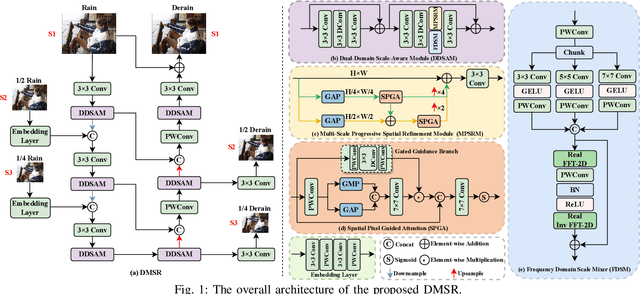
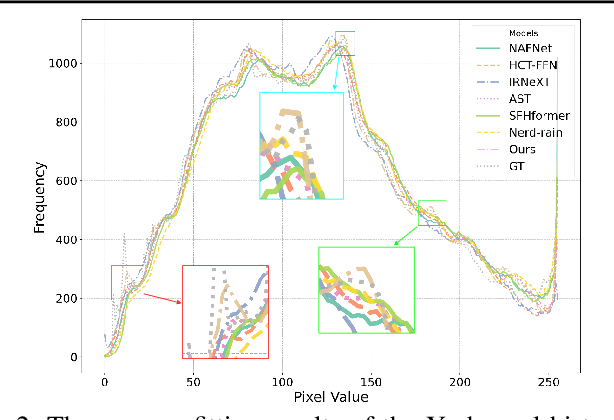
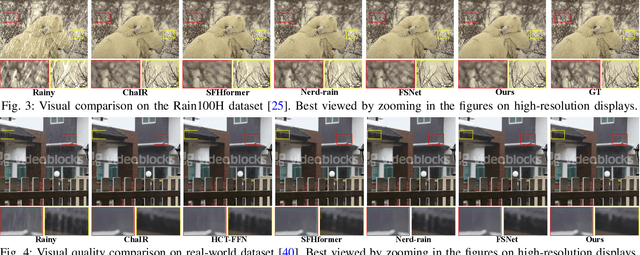
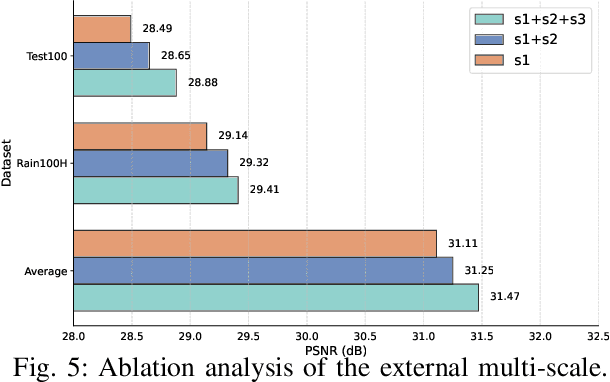
Abstract:Existing image deraining methods typically rely on single-input, single-output, and single-scale architectures, which overlook the joint multi-scale information between external and internal features. Furthermore, single-domain representations are often too restrictive, limiting their ability to handle the complexities of real-world rain scenarios. To address these challenges, we propose a novel Dual-Domain Multi-Scale Representation Network (DMSR). The key idea is to exploit joint multi-scale representations from both external and internal domains in parallel while leveraging the strengths of both spatial and frequency domains to capture more comprehensive properties. Specifically, our method consists of two main components: the Multi-Scale Progressive Spatial Refinement Module (MPSRM) and the Frequency Domain Scale Mixer (FDSM). The MPSRM enables the interaction and coupling of multi-scale expert information within the internal domain using a hierarchical modulation and fusion strategy. The FDSM extracts multi-scale local information in the spatial domain, while also modeling global dependencies in the frequency domain. Extensive experiments show that our model achieves state-of-the-art performance across six benchmark datasets.
Fraesormer: Learning Adaptive Sparse Transformer for Efficient Food Recognition
Mar 15, 2025Abstract:In recent years, Transformer has witnessed significant progress in food recognition. However, most existing approaches still face two critical challenges in lightweight food recognition: (1) the quadratic complexity and redundant feature representation from interactions with irrelevant tokens; (2) static feature recognition and single-scale representation, which overlook the unstructured, non-fixed nature of food images and the need for multi-scale features. To address these, we propose an adaptive and efficient sparse Transformer architecture (Fraesormer) with two core designs: Adaptive Top-k Sparse Partial Attention (ATK-SPA) and Hierarchical Scale-Sensitive Feature Gating Network (HSSFGN). ATK-SPA uses a learnable Gated Dynamic Top-K Operator (GDTKO) to retain critical attention scores, filtering low query-key matches that hinder feature aggregation. It also introduces a partial channel mechanism to reduce redundancy and promote expert information flow, enabling local-global collaborative modeling. HSSFGN employs gating mechanism to achieve multi-scale feature representation, enhancing contextual semantic information. Extensive experiments show that Fraesormer outperforms state-of-the-art methods. code is available at https://zs1314.github.io/Fraesormer.
SkinMamba: A Precision Skin Lesion Segmentation Architecture with Cross-Scale Global State Modeling and Frequency Boundary Guidance
Sep 17, 2024



Abstract:Skin lesion segmentation is a crucial method for identifying early skin cancer. In recent years, both convolutional neural network (CNN) and Transformer-based methods have been widely applied. Moreover, combining CNN and Transformer effectively integrates global and local relationships, but remains limited by the quadratic complexity of Transformer. To address this, we propose a hybrid architecture based on Mamba and CNN, called SkinMamba. It maintains linear complexity while offering powerful long-range dependency modeling and local feature extraction capabilities. Specifically, we introduce the Scale Residual State Space Block (SRSSB), which captures global contextual relationships and cross-scale information exchange at a macro level, enabling expert communication in a global state. This effectively addresses challenges in skin lesion segmentation related to varying lesion sizes and inconspicuous target areas. Additionally, to mitigate boundary blurring and information loss during model downsampling, we introduce the Frequency Boundary Guided Module (FBGM), providing sufficient boundary priors to guide precise boundary segmentation, while also using the retained information to assist the decoder in the decoding process. Finally, we conducted comparative and ablation experiments on two public lesion segmentation datasets (ISIC2017 and ISIC2018), and the results demonstrate the strong competitiveness of SkinMamba in skin lesion segmentation tasks. The code is available at https://github.com/zs1314/SkinMamba.
OCTAMamba: A State-Space Model Approach for Precision OCTA Vasculature Segmentation
Sep 12, 2024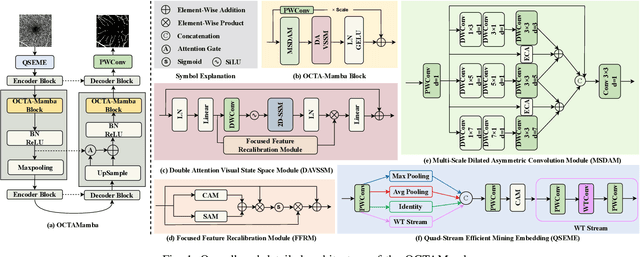
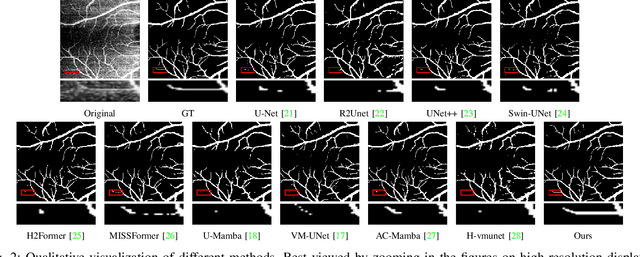
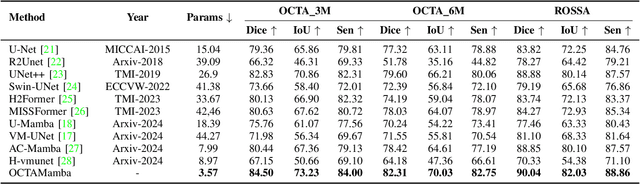
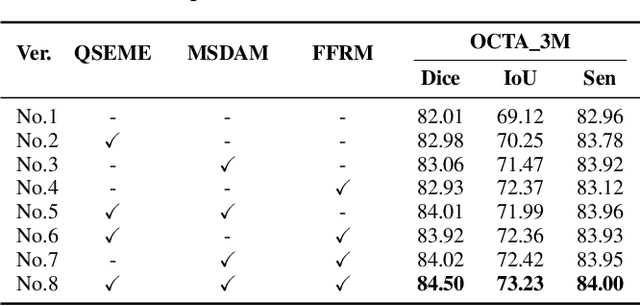
Abstract:Optical Coherence Tomography Angiography (OCTA) is a crucial imaging technique for visualizing retinal vasculature and diagnosing eye diseases such as diabetic retinopathy and glaucoma. However, precise segmentation of OCTA vasculature remains challenging due to the multi-scale vessel structures and noise from poor image quality and eye lesions. In this study, we proposed OCTAMamba, a novel U-shaped network based on the Mamba architecture, designed to segment vasculature in OCTA accurately. OCTAMamba integrates a Quad Stream Efficient Mining Embedding Module for local feature extraction, a Multi-Scale Dilated Asymmetric Convolution Module to capture multi-scale vasculature, and a Focused Feature Recalibration Module to filter noise and highlight target areas. Our method achieves efficient global modeling and local feature extraction while maintaining linear complexity, making it suitable for low-computation medical applications. Extensive experiments on the OCTA 3M, OCTA 6M, and ROSSA datasets demonstrated that OCTAMamba outperforms state-of-the-art methods, providing a new reference for efficient OCTA segmentation. Code is available at https://github.com/zs1314/OCTAMamba
Microscopic-Mamba: Revealing the Secrets of Microscopic Images with Just 4M Parameters
Sep 12, 2024



Abstract:In the field of medical microscopic image classification (MIC), CNN-based and Transformer-based models have been extensively studied. However, CNNs struggle with modeling long-range dependencies, limiting their ability to fully utilize semantic information in images. Conversely, Transformers are hampered by the complexity of quadratic computations. To address these challenges, we propose a model based on the Mamba architecture: Microscopic-Mamba. Specifically, we designed the Partially Selected Feed-Forward Network (PSFFN) to replace the last linear layer of the Visual State Space Module (VSSM), enhancing Mamba's local feature extraction capabilities. Additionally, we introduced the Modulation Interaction Feature Aggregation (MIFA) module to effectively modulate and dynamically aggregate global and local features. We also incorporated a parallel VSSM mechanism to improve inter-channel information interaction while reducing the number of parameters. Extensive experiments have demonstrated that our method achieves state-of-the-art performance on five public datasets. Code is available at https://github.com/zs1314/Microscopic-Mamba
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge