Mengyun Chen
Few-Shot Learning of Accurate Folding Landscape for Protein Structure Prediction
Aug 20, 2022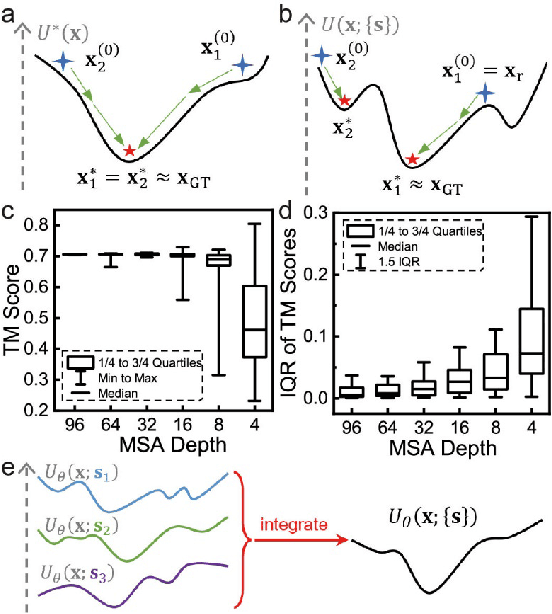
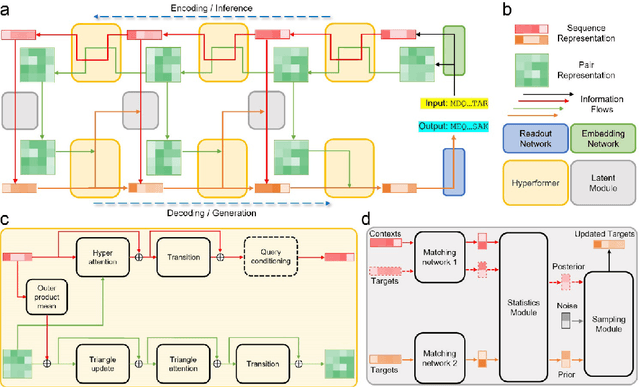
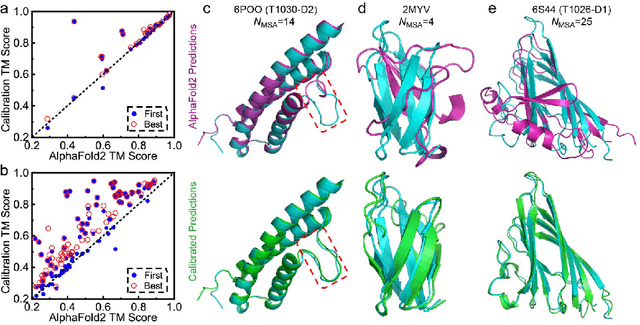
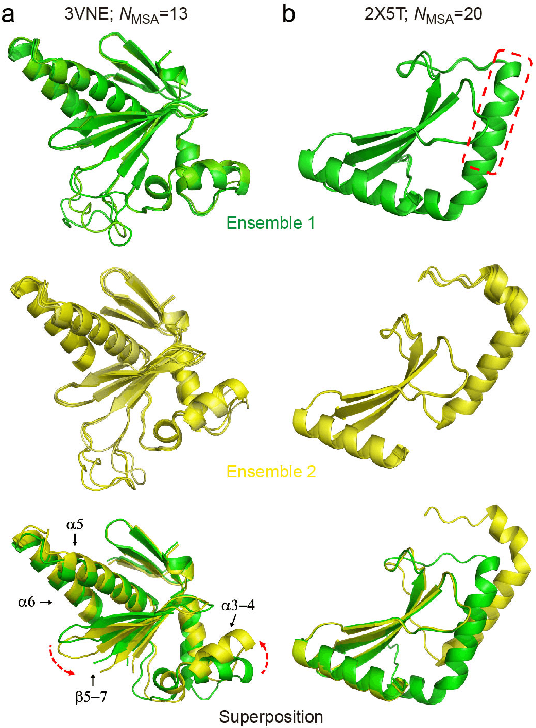
Abstract:Data-driven predictive methods which can efficiently and accurately transform protein sequences into biologically active structures are highly valuable for scientific research and therapeutical development. Determining accurate folding landscape using co-evolutionary information is fundamental to the success of modern protein structure prediction methods. As the state of the art, AlphaFold2 has dramatically raised the accuracy without performing explicit co-evolutionary analysis. Nevertheless, its performance still shows strong dependence on available sequence homologs. We investigated the cause of such dependence and presented EvoGen, a meta generative model, to remedy the underperformance of AlphaFold2 for poor MSA targets. EvoGen allows us to manipulate the folding landscape either by denoising the searched MSA or by generating virtual MSA, and helps AlphaFold2 fold accurately in low-data regime or even achieve encouraging performance with single-sequence predictions. Being able to make accurate predictions with few-shot MSA not only generalizes AlphaFold2 better for orphan sequences, but also democratizes its use for high-throughput applications. Besides, EvoGen combined with AlphaFold2 yields a probabilistic structure generation method which could explore alternative conformations of protein sequences, and the task-aware differentiable algorithm for sequence generation will benefit other related tasks including protein design.
PSP: Million-level Protein Sequence Dataset for Protein Structure Prediction
Jun 24, 2022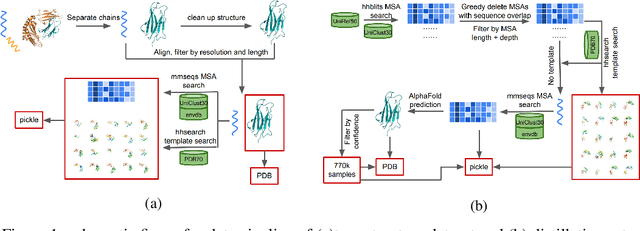
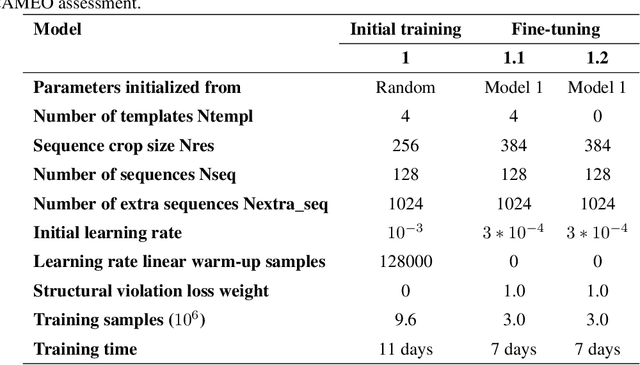
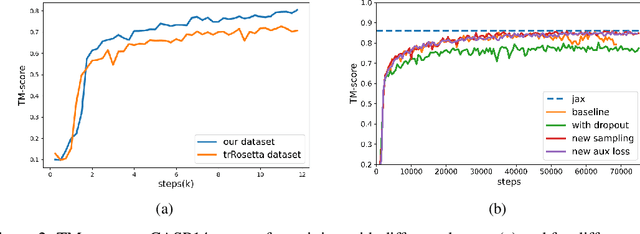
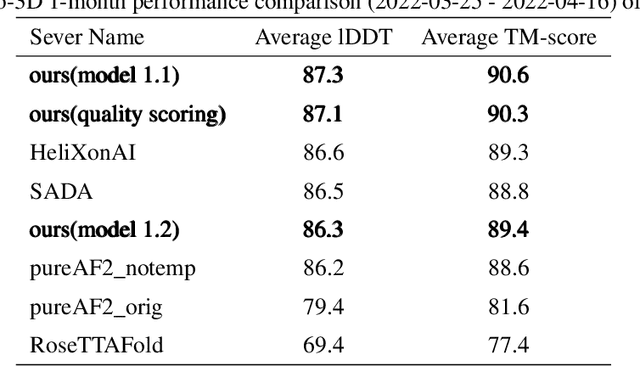
Abstract:Proteins are essential component of human life and their structures are important for function and mechanism analysis. Recent work has shown the potential of AI-driven methods for protein structure prediction. However, the development of new models is restricted by the lack of dataset and benchmark training procedure. To the best of our knowledge, the existing open source datasets are far less to satisfy the needs of modern protein sequence-structure related research. To solve this problem, we present the first million-level protein structure prediction dataset with high coverage and diversity, named as PSP. This dataset consists of 570k true structure sequences (10TB) and 745k complementary distillation sequences (15TB). We provide in addition the benchmark training procedure for SOTA protein structure prediction model on this dataset. We validate the utility of this dataset for training by participating CAMEO contest in which our model won the first place. We hope our PSP dataset together with the training benchmark can enable a broader community of AI/biology researchers for AI-driven protein related research.
Improving the Efficiency of Grammatical Error Correction with Erroneous Span Detection and Correction
Oct 07, 2020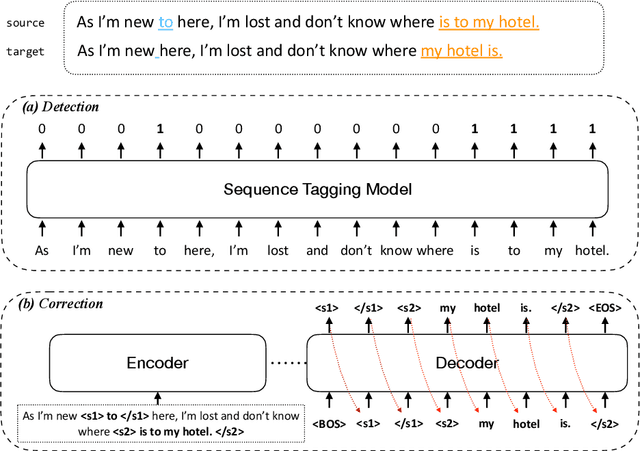
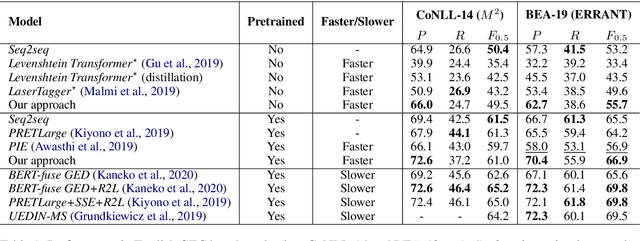
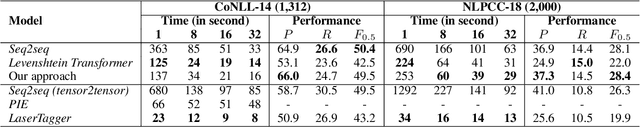
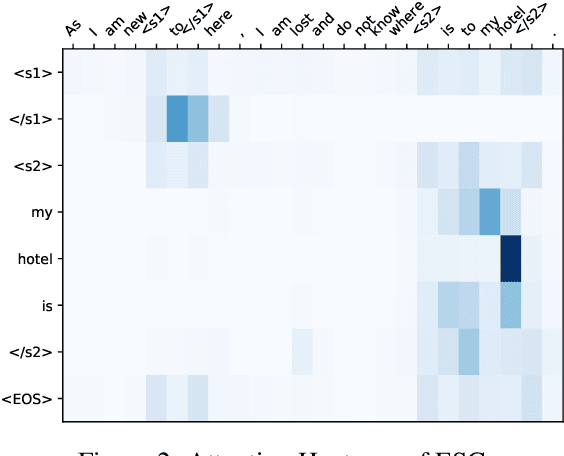
Abstract:We propose a novel language-independent approach to improve the efficiency for Grammatical Error Correction (GEC) by dividing the task into two subtasks: Erroneous Span Detection (ESD) and Erroneous Span Correction (ESC). ESD identifies grammatically incorrect text spans with an efficient sequence tagging model. Then, ESC leverages a seq2seq model to take the sentence with annotated erroneous spans as input and only outputs the corrected text for these spans. Experiments show our approach performs comparably to conventional seq2seq approaches in both English and Chinese GEC benchmarks with less than 50% time cost for inference.
Structured Stochastic Quasi-Newton Methods for Large-Scale Optimization Problems
Jun 17, 2020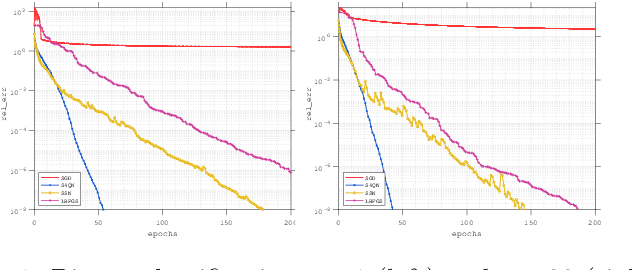
Abstract:In this paper, we consider large-scale finite-sum nonconvex problems arising from machine learning. Since the Hessian is often a summation of a relative cheap and accessible part and an expensive or even inaccessible part, a stochastic quasi-Newton matrix is constructed using partial Hessian information as much as possible. By further exploiting the low-rank structures based on the Nystr\"om approximation, the computation of the quasi-Newton direction is affordable. To make full use of the gradient estimation, we also develop an extra-step strategy for this framework. Global convergence to stationary point in expectation and local suplinear convergence rate are established under some mild assumptions. Numerical experiments on logistic regression, deep autoencoder networks and deep learning problems show that the efficiency of our proposed method is at least comparable with the state-of-the-art methods.
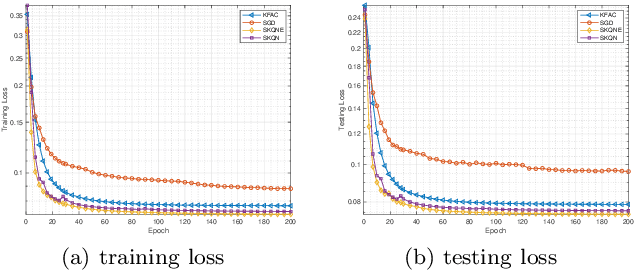
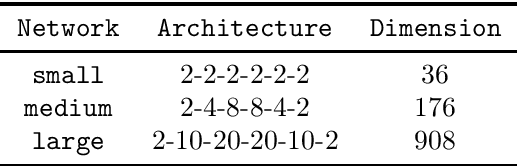
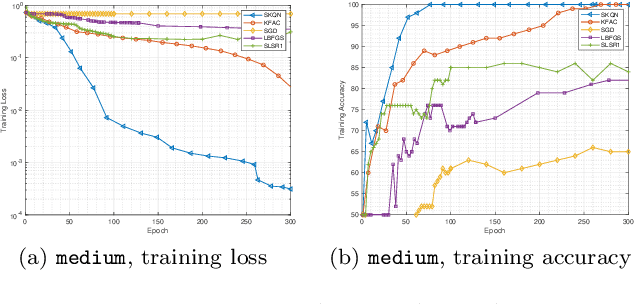
Sketchy Empirical Natural Gradient Methods for Deep Learning
Jun 16, 2020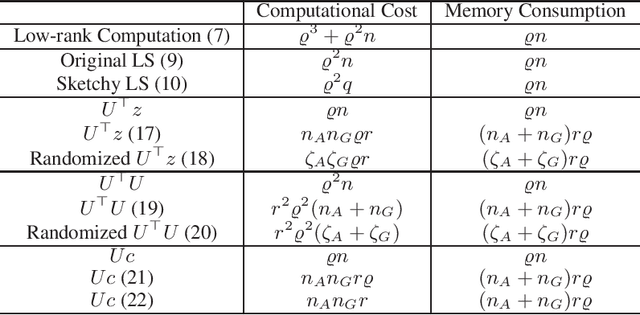
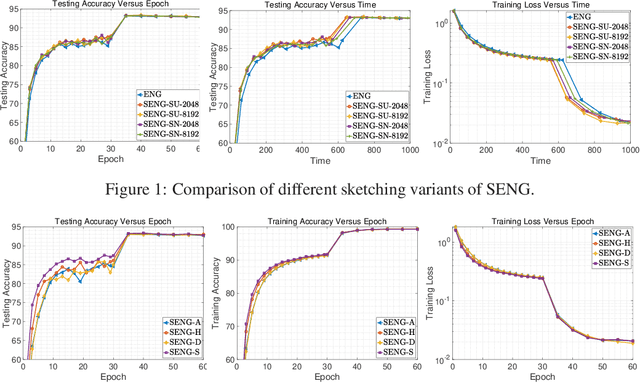
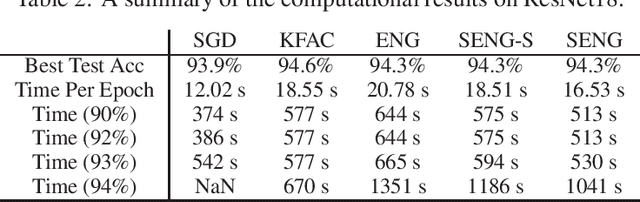
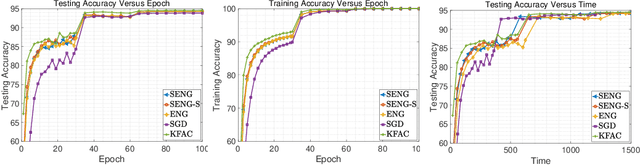
Abstract:In this paper, we develop an efficient sketchy empirical natural gradient method for large-scale finite-sum optimization problems from deep learning. The empirical Fisher information matrix is usually low-rank since the sampling is only practical on a small amount of data at each iteration. Although the corresponding natural gradient direction lies in a small subspace, both the computational cost and memory requirement are still not tractable due to the curse of dimensionality. We design randomized techniques for different neural network structures to resolve these challenges. For layers with a reasonable dimension, a sketching can be performed on a regularized least squares subproblem. Otherwise, since the gradient is a vectorization of the product between two matrices, we apply sketching on low-rank approximation of these matrices to compute the most expensive parts. Global convergence to stationary point is established under some mild assumptions. Numerical results on deep convolution networks illustrate that our method is quite competitive to the state-of-the-art methods such as SGD and KFAC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge