Matthew J. Thompson
Static Segmentation by Tracking: A Frustratingly Label-Efficient Approach to Fine-Grained Segmentation
Jan 12, 2025



Abstract:We study image segmentation in the biological domain, particularly trait and part segmentation from specimen images (e.g., butterfly wing stripes or beetle body parts). This is a crucial, fine-grained task that aids in understanding the biology of organisms. The conventional approach involves hand-labeling masks, often for hundreds of images per species, and training a segmentation model to generalize these labels to other images, which can be exceedingly laborious. We present a label-efficient method named Static Segmentation by Tracking (SST). SST is built upon the insight: while specimens of the same species have inherent variations, the traits and parts we aim to segment show up consistently. This motivates us to concatenate specimen images into a ``pseudo-video'' and reframe trait and part segmentation as a tracking problem. Concretely, SST generates masks for unlabeled images by propagating annotated or predicted masks from the ``pseudo-preceding'' images. Powered by Segment Anything Model 2 (SAM~2) initially developed for video segmentation, we show that SST can achieve high-quality trait and part segmentation with merely one labeled image per species -- a breakthrough for analyzing specimen images. We further develop a cycle-consistent loss to fine-tune the model, again using one labeled image. Additionally, we highlight the broader potential of SST, including one-shot instance segmentation on images taken in the wild and trait-based image retrieval.
VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological Images
Aug 28, 2024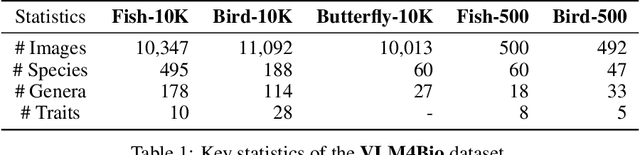
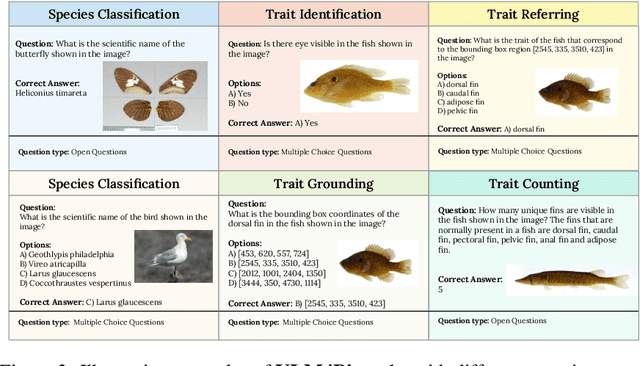

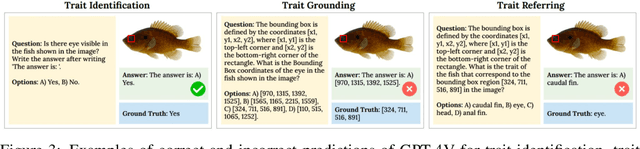
Abstract:Images are increasingly becoming the currency for documenting biodiversity on the planet, providing novel opportunities for accelerating scientific discoveries in the field of organismal biology, especially with the advent of large vision-language models (VLMs). We ask if pre-trained VLMs can aid scientists in answering a range of biologically relevant questions without any additional fine-tuning. In this paper, we evaluate the effectiveness of 12 state-of-the-art (SOTA) VLMs in the field of organismal biology using a novel dataset, VLM4Bio, consisting of 469K question-answer pairs involving 30K images from three groups of organisms: fishes, birds, and butterflies, covering five biologically relevant tasks. We also explore the effects of applying prompting techniques and tests for reasoning hallucination on the performance of VLMs, shedding new light on the capabilities of current SOTA VLMs in answering biologically relevant questions using images. The code and datasets for running all the analyses reported in this paper can be found at https://github.com/sammarfy/VLM4Bio.
Smartphone Camera Oximetry in an Induced Hypoxemia Study
Mar 31, 2021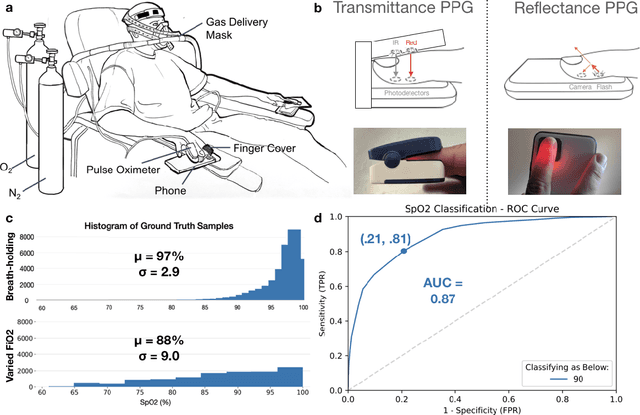
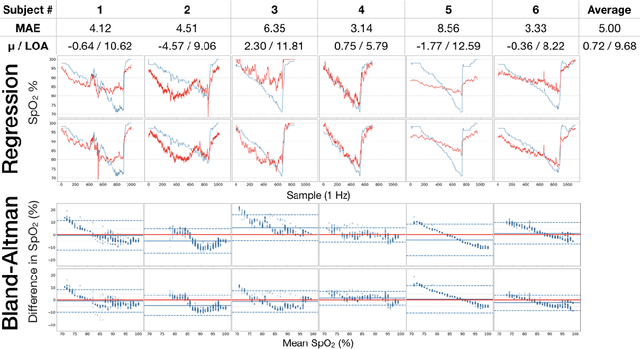
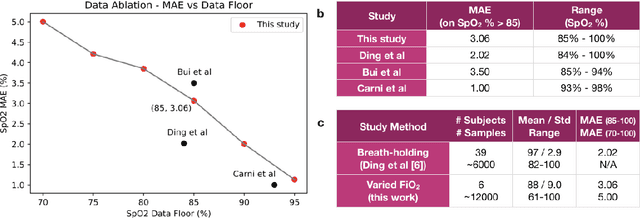
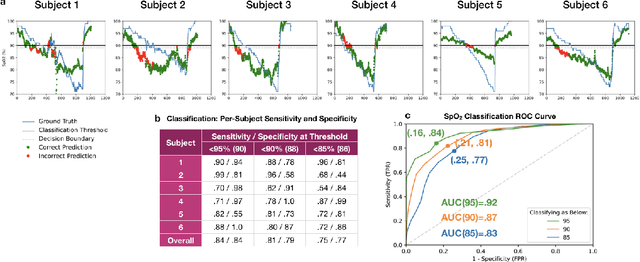
Abstract:Hypoxemia, a medical condition that occurs when the blood is not carrying enough oxygen to adequately supply the tissues, is a leading indicator for dangerous complications of respiratory diseases like asthma, COPD, and COVID-19. While purpose-built pulse oximeters can provide accurate blood-oxygen saturation (SpO$_2$) readings that allow for diagnosis of hypoxemia, enabling this capability in unmodified smartphone cameras via a software update could give more people access to important information about their health, as well as improve physicians' ability to remotely diagnose and treat respiratory conditions. In this work, we take a step towards this goal by performing the first clinical development validation on a smartphone-based SpO$_2$ sensing system using a varied fraction of inspired oxygen (FiO$_2$) protocol, creating a clinically relevant validation dataset for solely smartphone-based methods on a wide range of SpO$_2$ values (70%-100%) for the first time. This contrasts with previous studies, which evaluated performance on a far smaller range (85%-100%). We build a deep learning model using this data to demonstrate accurate reporting of SpO$_2$ level with an overall MAE=5.00% SpO$_2$ and identifying positive cases of low SpO$_2$<90% with 81% sensitivity and 79% specificity. We ground our analysis with a summary of recent literature in smartphone-based SpO2 monitoring, and we provide the data from the FiO$_2$ study in open-source format, so that others may build on this work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge