Shwetak N. Patel
Lifestyle-Informed Personalized Blood Biomarker Prediction via Novel Representation Learning
Jul 09, 2024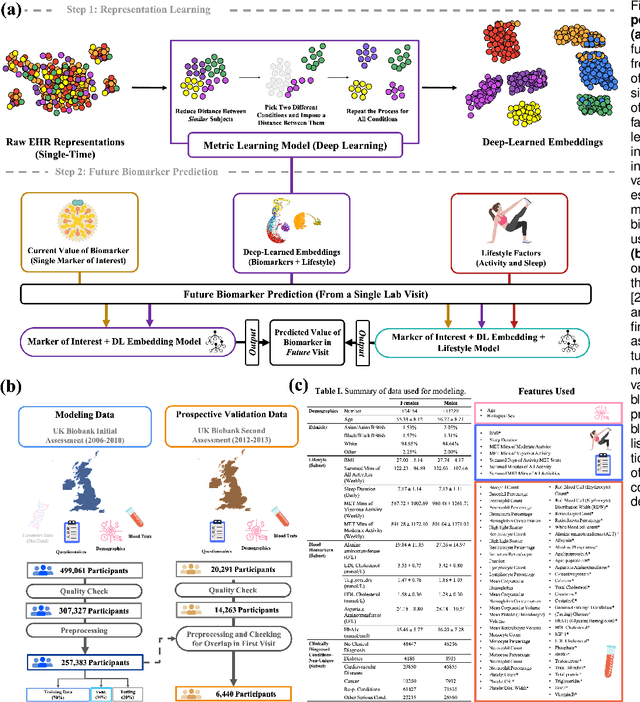
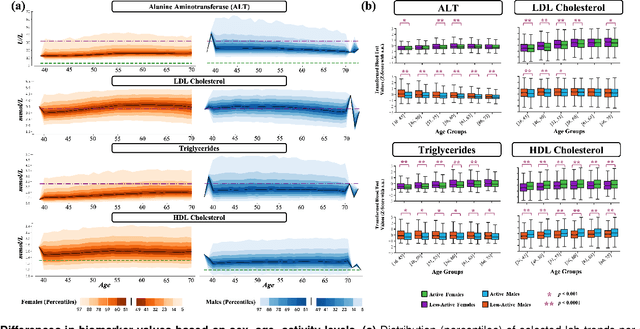
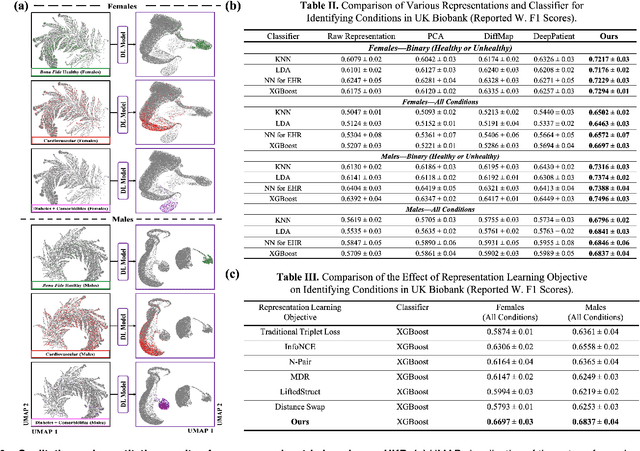
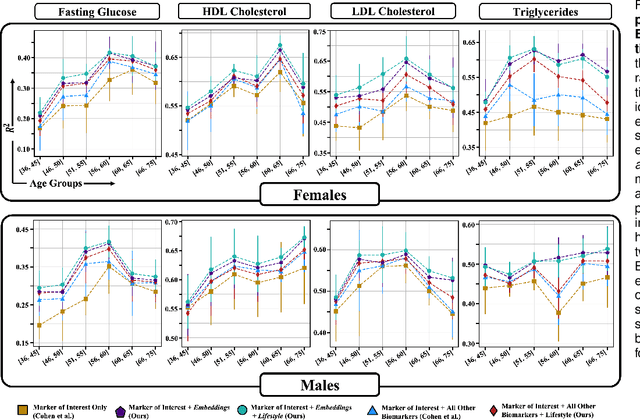
Abstract:Blood biomarkers are an essential tool for healthcare providers to diagnose, monitor, and treat a wide range of medical conditions. Current reference values and recommended ranges often rely on population-level statistics, which may not adequately account for the influence of inter-individual variability driven by factors such as lifestyle and genetics. In this work, we introduce a novel framework for predicting future blood biomarker values and define personalized references through learned representations from lifestyle data (physical activity and sleep) and blood biomarkers. Our proposed method learns a similarity-based embedding space that captures the complex relationship between biomarkers and lifestyle factors. Using the UK Biobank (257K participants), our results show that our deep-learned embeddings outperform traditional and current state-of-the-art representation learning techniques in predicting clinical diagnosis. Using a subset of UK Biobank of 6440 participants who have follow-up visits, we validate that the inclusion of these embeddings and lifestyle factors directly in blood biomarker models improves the prediction of future lab values from a single lab visit. This personalized modeling approach provides a foundation for developing more accurate risk stratification tools and tailoring preventative care strategies. In clinical settings, this translates to the potential for earlier disease detection, more timely interventions, and ultimately, a shift towards personalized healthcare.
Smartphone Camera Oximetry in an Induced Hypoxemia Study
Mar 31, 2021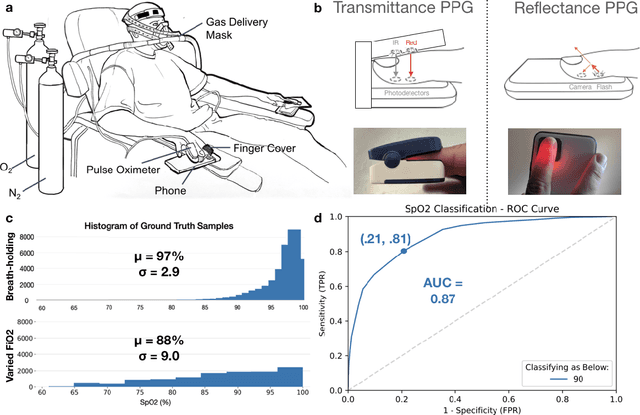
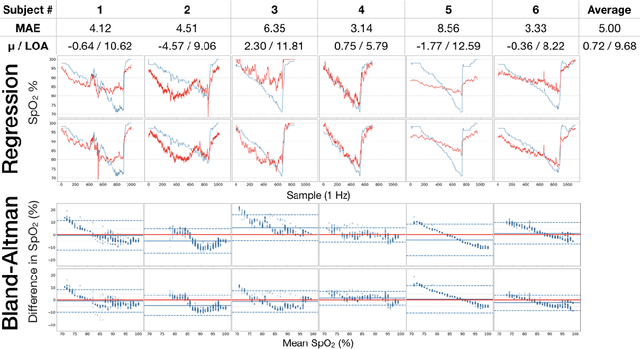
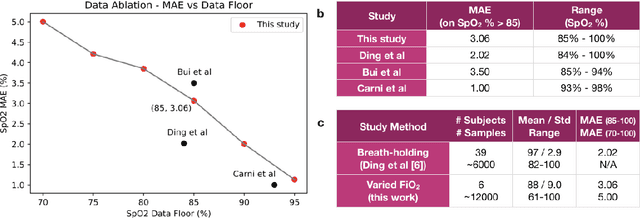
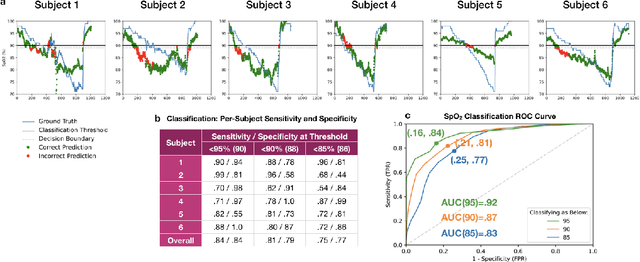
Abstract:Hypoxemia, a medical condition that occurs when the blood is not carrying enough oxygen to adequately supply the tissues, is a leading indicator for dangerous complications of respiratory diseases like asthma, COPD, and COVID-19. While purpose-built pulse oximeters can provide accurate blood-oxygen saturation (SpO$_2$) readings that allow for diagnosis of hypoxemia, enabling this capability in unmodified smartphone cameras via a software update could give more people access to important information about their health, as well as improve physicians' ability to remotely diagnose and treat respiratory conditions. In this work, we take a step towards this goal by performing the first clinical development validation on a smartphone-based SpO$_2$ sensing system using a varied fraction of inspired oxygen (FiO$_2$) protocol, creating a clinically relevant validation dataset for solely smartphone-based methods on a wide range of SpO$_2$ values (70%-100%) for the first time. This contrasts with previous studies, which evaluated performance on a far smaller range (85%-100%). We build a deep learning model using this data to demonstrate accurate reporting of SpO$_2$ level with an overall MAE=5.00% SpO$_2$ and identifying positive cases of low SpO$_2$<90% with 81% sensitivity and 79% specificity. We ground our analysis with a summary of recent literature in smartphone-based SpO2 monitoring, and we provide the data from the FiO$_2$ study in open-source format, so that others may build on this work.
Transfer Learning for Activity Recognition in Mobile Health
Jul 12, 2020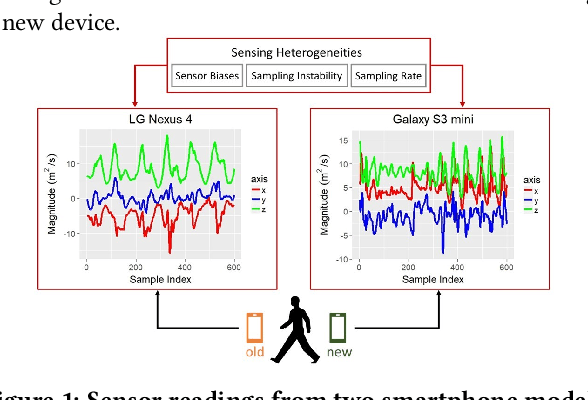
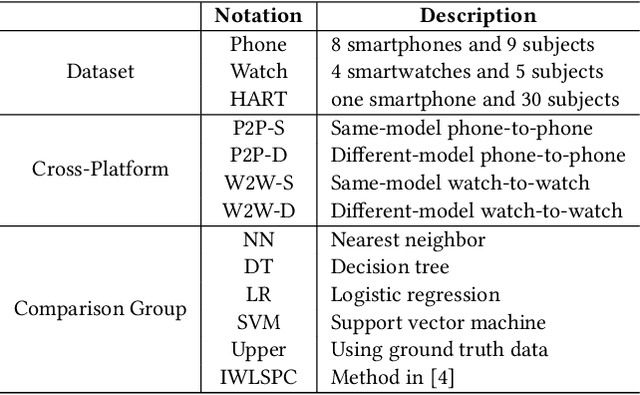
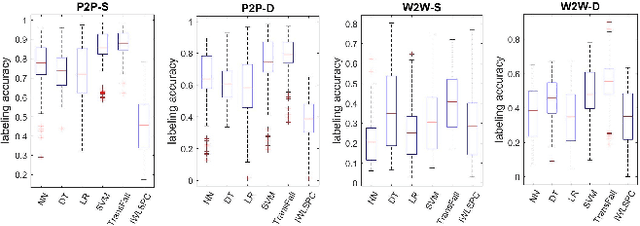
Abstract:While activity recognition from inertial sensors holds potential for mobile health, differences in sensing platforms and user movement patterns cause performance degradation. Aiming to address these challenges, we propose a transfer learning framework, TransFall, for sensor-based activity recognition. TransFall's design contains a two-tier data transformation, a label estimation layer, and a model generation layer to recognize activities for the new scenario. We validate TransFall analytically and empirically.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge