Donna Spruijt-Metz
Center of Economic and Social Research, University of Southern California, Los Angeles, California, U.S.A
BayesLDM: A Domain-Specific Language for Probabilistic Modeling of Longitudinal Data
Sep 12, 2022


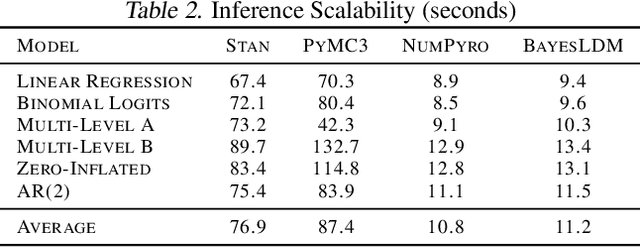
Abstract:In this paper we present BayesLDM, a system for Bayesian longitudinal data modeling consisting of a high-level modeling language with specific features for modeling complex multivariate time series data coupled with a compiler that can produce optimized probabilistic program code for performing inference in the specified model. BayesLDM supports modeling of Bayesian network models with a specific focus on the efficient, declarative specification of dynamic Bayesian Networks (DBNs). The BayesLDM compiler combines a model specification with inspection of available data and outputs code for performing Bayesian inference for unknown model parameters while simultaneously handling missing data. These capabilities have the potential to significantly accelerate iterative modeling workflows in domains that involve the analysis of complex longitudinal data by abstracting away the process of producing computationally efficient probabilistic inference code. We describe the BayesLDM system components, evaluate the efficiency of representation and inference optimizations and provide an illustrative example of the application of the system to analyzing heterogeneous and partially observed mobile health data.
Transfer Learning for Activity Recognition in Mobile Health
Jul 12, 2020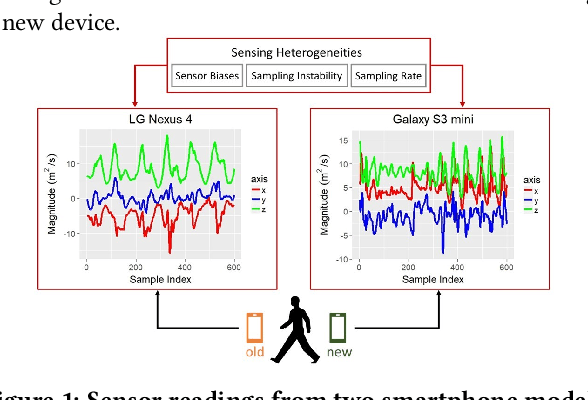
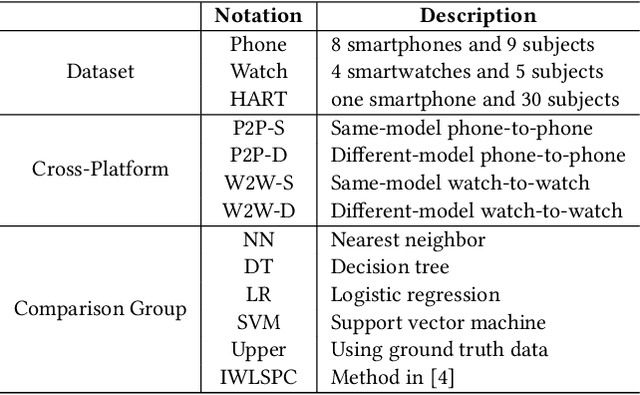
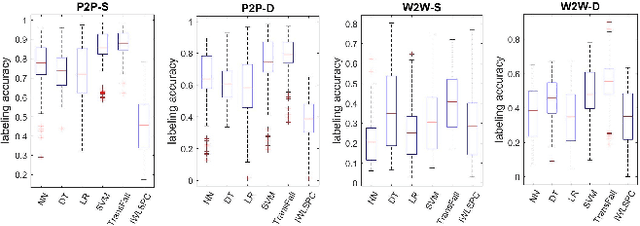
Abstract:While activity recognition from inertial sensors holds potential for mobile health, differences in sensing platforms and user movement patterns cause performance degradation. Aiming to address these challenges, we propose a transfer learning framework, TransFall, for sensor-based activity recognition. TransFall's design contains a two-tier data transformation, a label estimation layer, and a model generation layer to recognize activities for the new scenario. We validate TransFall analytically and empirically.
Estimating Individualized Treatment Regimes from Crossover Designs
Feb 05, 2019



Abstract:The field of precision medicine aims to tailor treatment based on patient-specific factors in a reproducible way. To this end, estimating an optimal individualized treatment regime (ITR) that recommends treatment decisions based on patient characteristics to maximize the mean of a pre-specified outcome is of particular interest. Several methods have been proposed for estimating an optimal ITR from clinical trial data in the parallel group setting where each subject is randomized to a single intervention. However, little work has been done in the area of estimating the optimal ITR from crossover study designs. Such designs naturally lend themselves to precision medicine, because they allow for observing the response to multiple treatments for each patient. In this paper, we introduce a method for estimating the optimal ITR using data from a 2x2 crossover study with or without carryover effects. The proposed method is similar to policy search methods such as outcome weighted learning; however, we take advantage of the crossover design by using the difference in responses under each treatment as the observed reward. We establish Fisher and global consistency, present numerical experiments, and analyze data from a feeding trial to demonstrate the improved performance of the proposed method compared to standard methods for a parallel study design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge