Daniel Rivera
Artificial intelligence application in lymphoma diagnosis: from Convolutional Neural Network to Vision Transformer
Apr 05, 2025Abstract:Recently, vision transformers were shown to be capable of outperforming convolutional neural networks when pretrained on sufficiently large datasets. Vision transformer models show good accuracy on large scale datasets, with features of multi-modal training. Due to their promising feature detection, we aim to explore vision transformer models for diagnosis of anaplastic large cell lymphoma versus classical Hodgkin lymphoma using pathology whole slide images of HE slides. We compared the classification performance of the vision transformer to our previously designed convolutional neural network on the same dataset. The dataset includes whole slide images of HE slides for 20 cases, including 10 cases in each diagnostic category. From each whole slide image, 60 image patches having size of 100 by 100 pixels and at magnification of 20 were obtained to yield 1200 image patches, from which 90 percent were used for training, 9 percent for validation, and 10 percent for testing. The test results from the convolutional neural network model had previously shown an excellent diagnostic accuracy of 100 percent. The test results from the vision transformer model also showed a comparable accuracy at 100 percent. To the best of the authors' knowledge, this is the first direct comparison of predictive performance between a vision transformer model and a convolutional neural network model using the same dataset of lymphoma. Overall, convolutional neural network has a more mature architecture than vision transformer and is usually the best choice when large scale pretraining is not an available option. Nevertheless, our current study shows comparable and excellent accuracy of vision transformer compared to that of convolutional neural network even with a relatively small dataset of anaplastic large cell lymphoma and classical Hodgkin lymphoma.
BayesLDM: A Domain-Specific Language for Probabilistic Modeling of Longitudinal Data
Sep 12, 2022


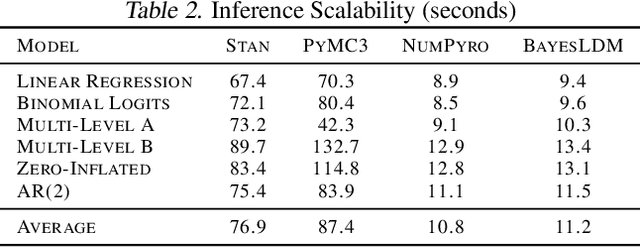
Abstract:In this paper we present BayesLDM, a system for Bayesian longitudinal data modeling consisting of a high-level modeling language with specific features for modeling complex multivariate time series data coupled with a compiler that can produce optimized probabilistic program code for performing inference in the specified model. BayesLDM supports modeling of Bayesian network models with a specific focus on the efficient, declarative specification of dynamic Bayesian Networks (DBNs). The BayesLDM compiler combines a model specification with inspection of available data and outputs code for performing Bayesian inference for unknown model parameters while simultaneously handling missing data. These capabilities have the potential to significantly accelerate iterative modeling workflows in domains that involve the analysis of complex longitudinal data by abstracting away the process of producing computationally efficient probabilistic inference code. We describe the BayesLDM system components, evaluate the efficiency of representation and inference optimizations and provide an illustrative example of the application of the system to analyzing heterogeneous and partially observed mobile health data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge