Amer Wahed
Artificial intelligence application in lymphoma diagnosis: from Convolutional Neural Network to Vision Transformer
Apr 05, 2025Abstract:Recently, vision transformers were shown to be capable of outperforming convolutional neural networks when pretrained on sufficiently large datasets. Vision transformer models show good accuracy on large scale datasets, with features of multi-modal training. Due to their promising feature detection, we aim to explore vision transformer models for diagnosis of anaplastic large cell lymphoma versus classical Hodgkin lymphoma using pathology whole slide images of HE slides. We compared the classification performance of the vision transformer to our previously designed convolutional neural network on the same dataset. The dataset includes whole slide images of HE slides for 20 cases, including 10 cases in each diagnostic category. From each whole slide image, 60 image patches having size of 100 by 100 pixels and at magnification of 20 were obtained to yield 1200 image patches, from which 90 percent were used for training, 9 percent for validation, and 10 percent for testing. The test results from the convolutional neural network model had previously shown an excellent diagnostic accuracy of 100 percent. The test results from the vision transformer model also showed a comparable accuracy at 100 percent. To the best of the authors' knowledge, this is the first direct comparison of predictive performance between a vision transformer model and a convolutional neural network model using the same dataset of lymphoma. Overall, convolutional neural network has a more mature architecture than vision transformer and is usually the best choice when large scale pretraining is not an available option. Nevertheless, our current study shows comparable and excellent accuracy of vision transformer compared to that of convolutional neural network even with a relatively small dataset of anaplastic large cell lymphoma and classical Hodgkin lymphoma.
Deep Learning Provides Rapid Screen for Breast Cancer Metastasis with Sentinel Lymph Nodes
Jan 14, 2023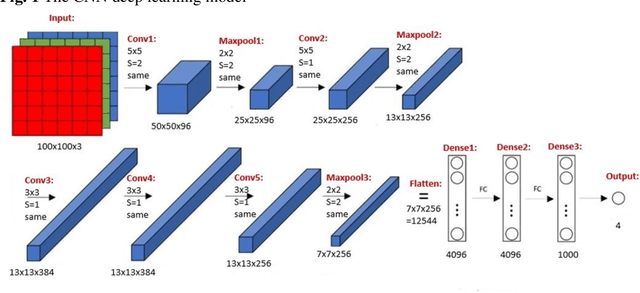
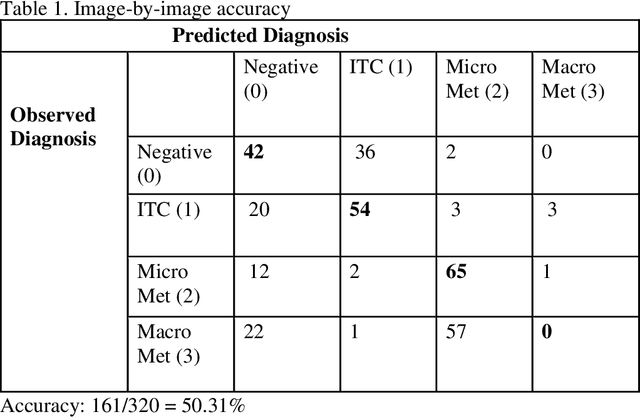
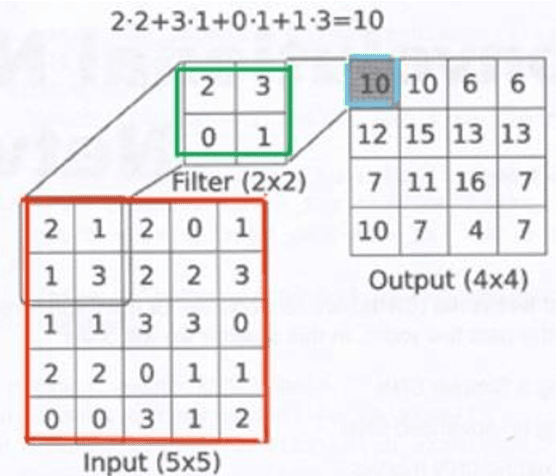
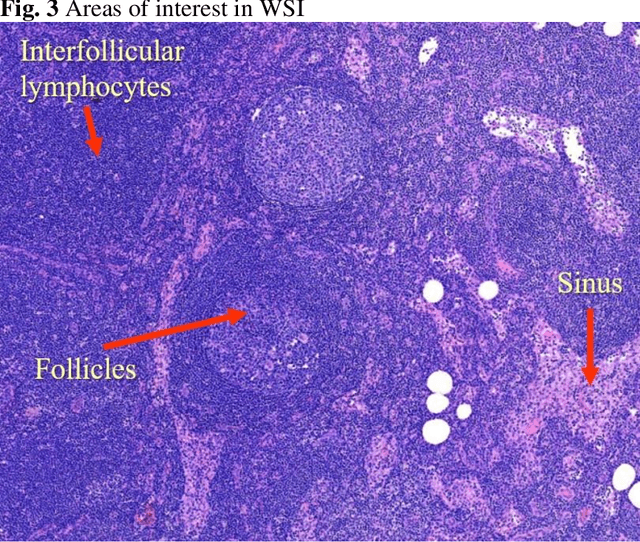
Abstract:Deep learning has been shown to be useful to detect breast cancer metastases by analyzing whole slide images of sentinel lymph nodes. However, it requires extensive scanning and analysis of all the lymph nodes slides for each case. Our deep learning study focuses on breast cancer screening with only a small set of image patches from any sentinel lymph node, positive or negative for metastasis, to detect changes in tumor environment and not in the tumor itself. We design a convolutional neural network in the Python language to build a diagnostic model for this purpose. The excellent results from this preliminary study provided a proof of concept for incorporating automated metastatic screen into the digital pathology workflow to augment the pathologists' productivity. Our approach is unique since it provides a very rapid screen rather than an exhaustive search for tumor in all fields of all sentinel lymph nodes.
Proteomics Analysis of FLT3-ITD Mutation in Acute Myeloid Leukemia Using Deep Learning Neural Network
Dec 29, 2017



Abstract:Deep Learning can significantly benefit cancer proteomics and genomics. In this study, we attempt to determine a set of critical proteins that are associated with the FLT3-ITD mutation in newly-diagnosed acute myeloid leukemia patients. A Deep Learning network consisting of autoencoders forming a hierarchical model from which high-level features are extracted without labeled training data. Dimensional reduction reduced the number of critical proteins from 231 to 20. Deep Learning found an excellent correlation between FLT3-ITD mutation with the levels of these 20 critical proteins (accuracy 97%, sensitivity 90%, specificity 100%). Our Deep Learning network could hone in on 20 proteins with the strongest association with FLT3-ITD. The results of this study allow a novel approach to determine critical protein pathways in the FLT3-ITD mutation, and provide proof-of-concept for an accurate approach to model big data in cancer proteomics and genomics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge