Hassan Ghasemzadeh
Counterfactual Modeling with Fine-Tuned LLMs for Health Intervention Design and Sensor Data Augmentation
Jan 21, 2026Abstract:Counterfactual explanations (CFEs) provide human-centric interpretability by identifying the minimal, actionable changes required to alter a machine learning model's prediction. Therefore, CFs can be used as (i) interventions for abnormality prevention and (ii) augmented data for training robust models. We conduct a comprehensive evaluation of CF generation using large language models (LLMs), including GPT-4 (zero-shot and few-shot) and two open-source models-BioMistral-7B and LLaMA-3.1-8B, in both pretrained and fine-tuned configurations. Using the multimodal AI-READI clinical dataset, we assess CFs across three dimensions: intervention quality, feature diversity, and augmentation effectiveness. Fine-tuned LLMs, particularly LLaMA-3.1-8B, produce CFs with high plausibility (up to 99%), strong validity (up to 0.99), and realistic, behaviorally modifiable feature adjustments. When used for data augmentation under controlled label-scarcity settings, LLM-generated CFs substantially restore classifier performance, yielding an average 20% F1 recovery across three scarcity scenarios. Compared with optimization-based baselines such as DiCE, CFNOW, and NICE, LLMs offer a flexible, model-agnostic approach that generates more clinically actionable and semantically coherent counterfactuals. Overall, this work demonstrates the promise of LLM-driven counterfactuals for both interpretable intervention design and data-efficient model training in sensor-based digital health. Impact: SenseCF fine-tunes an LLM to generate valid, representative counterfactual explanations and supplement minority class in an imbalanced dataset for improving model training and boosting model robustness and predictive performance
Guiding Diffusion with Deep Geometric Moments: Balancing Fidelity and Variation
May 18, 2025Abstract:Text-to-image generation models have achieved remarkable capabilities in synthesizing images, but often struggle to provide fine-grained control over the output. Existing guidance approaches, such as segmentation maps and depth maps, introduce spatial rigidity that restricts the inherent diversity of diffusion models. In this work, we introduce Deep Geometric Moments (DGM) as a novel form of guidance that encapsulates the subject's visual features and nuances through a learned geometric prior. DGMs focus specifically on the subject itself compared to DINO or CLIP features, which suffer from overemphasis on global image features or semantics. Unlike ResNets, which are sensitive to pixel-wise perturbations, DGMs rely on robust geometric moments. Our experiments demonstrate that DGM effectively balance control and diversity in diffusion-based image generation, allowing a flexible control mechanism for steering the diffusion process.
GlyTwin: Digital Twin for Glucose Control in Type 1 Diabetes Through Optimal Behavioral Modifications Using Patient-Centric Counterfactuals
Apr 14, 2025Abstract:Frequent and long-term exposure to hyperglycemia (i.e., high blood glucose) increases the risk of chronic complications such as neuropathy, nephropathy, and cardiovascular disease. Current technologies like continuous subcutaneous insulin infusion (CSII) and continuous glucose monitoring (CGM) primarily model specific aspects of glycemic control-like hypoglycemia prediction or insulin delivery. Similarly, most digital twin approaches in diabetes management simulate only physiological processes. These systems lack the ability to offer alternative treatment scenarios that support proactive behavioral interventions. To address this, we propose GlyTwin, a novel digital twin framework that uses counterfactual explanations to simulate optimal treatments for glucose regulation. Our approach helps patients and caregivers modify behaviors like carbohydrate intake and insulin dosing to avoid abnormal glucose events. GlyTwin generates behavioral treatment suggestions that proactively prevent hyperglycemia by recommending small adjustments to daily choices, reducing both frequency and duration of these events. Additionally, it incorporates stakeholder preferences into the intervention design, making recommendations patient-centric and tailored. We evaluate GlyTwin on AZT1D, a newly constructed dataset with longitudinal data from 21 type 1 diabetes (T1D) patients on automated insulin delivery systems over 26 days. Results show GlyTwin outperforms state-of-the-art counterfactual methods, generating 76.6% valid and 86% effective interventions. These findings demonstrate the promise of counterfactual-driven digital twins in delivering personalized healthcare.
Enhancing Metabolic Syndrome Prediction with Hybrid Data Balancing and Counterfactuals
Apr 09, 2025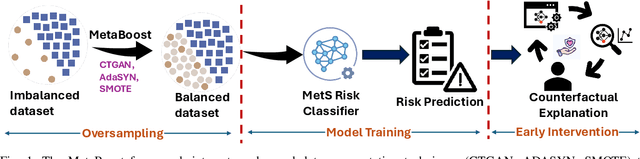

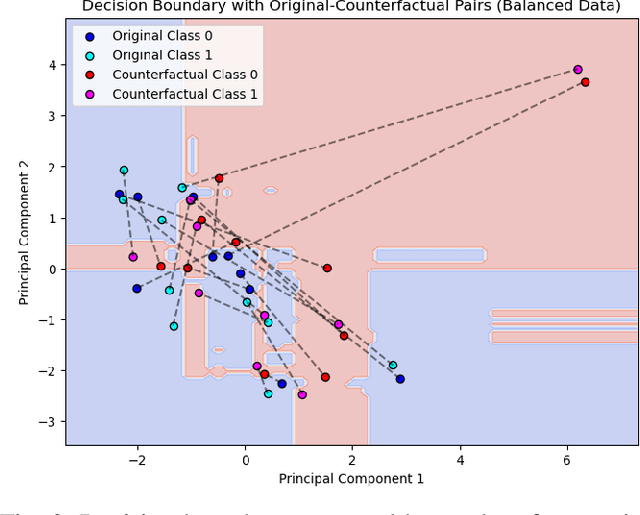

Abstract:Metabolic Syndrome (MetS) is a cluster of interrelated risk factors that significantly increases the risk of cardiovascular diseases and type 2 diabetes. Despite its global prevalence, accurate prediction of MetS remains challenging due to issues such as class imbalance, data scarcity, and methodological inconsistencies in existing studies. In this paper, we address these challenges by systematically evaluating and optimizing machine learning (ML) models for MetS prediction, leveraging advanced data balancing techniques and counterfactual analysis. Multiple ML models, including XGBoost, Random Forest, TabNet, etc., were trained and compared under various data balancing techniques such as random oversampling (ROS), SMOTE, ADASYN, and CTGAN. Additionally, we introduce MetaBoost, a novel hybrid framework that integrates SMOTE, ADASYN, and CTGAN, optimizing synthetic data generation through weighted averaging and iterative weight tuning to enhance the model's performance (achieving a 1.14% accuracy improvement over individual balancing techniques). A comprehensive counterfactual analysis is conducted to quantify feature-level changes required to shift individuals from high-risk to low-risk categories. The results indicate that blood glucose (50.3%) and triglycerides (46.7%) were the most frequently modified features, highlighting their clinical significance in MetS risk reduction. Additionally, probabilistic analysis shows elevated blood glucose (85.5% likelihood) and triglycerides (74.9% posterior probability) as the strongest predictors. This study not only advances the methodological rigor of MetS prediction but also provides actionable insights for clinicians and researchers, highlighting the potential of ML in mitigating the public health burden of metabolic syndrome.
AIMI: Leveraging Future Knowledge and Personalization in Sparse Event Forecasting for Treatment Adherence
Mar 20, 2025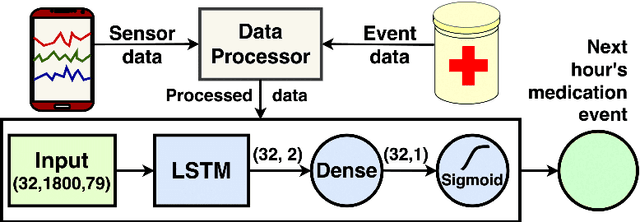

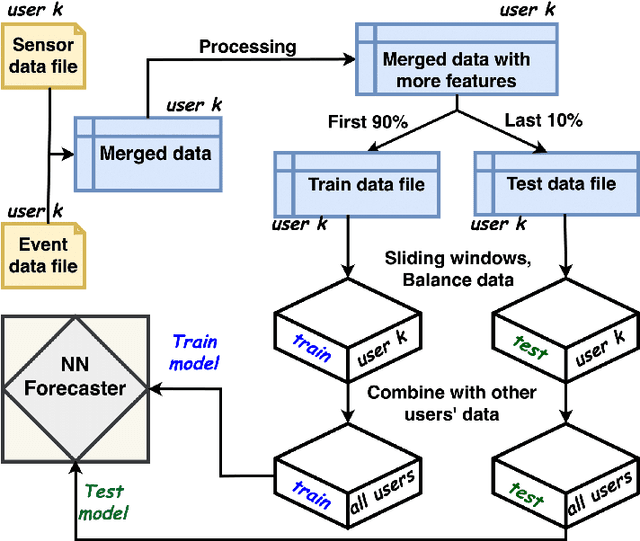

Abstract:Adherence to prescribed treatments is crucial for individuals with chronic conditions to avoid costly or adverse health outcomes. For certain patient groups, intensive lifestyle interventions are vital for enhancing medication adherence. Accurate forecasting of treatment adherence can open pathways to developing an on-demand intervention tool, enabling timely and personalized support. With the increasing popularity of smartphones and wearables, it is now easier than ever to develop and deploy smart activity monitoring systems. However, effective forecasting systems for treatment adherence based on wearable sensors are still not widely available. We close this gap by proposing Adherence Forecasting and Intervention with Machine Intelligence (AIMI). AIMI is a knowledge-guided adherence forecasting system that leverages smartphone sensors and previous medication history to estimate the likelihood of forgetting to take a prescribed medication. A user study was conducted with 27 participants who took daily medications to manage their cardiovascular diseases. We designed and developed CNN and LSTM-based forecasting models with various combinations of input features and found that LSTM models can forecast medication adherence with an accuracy of 0.932 and an F-1 score of 0.936. Moreover, through a series of ablation studies involving convolutional and recurrent neural network architectures, we demonstrate that leveraging known knowledge about future and personalized training enhances the accuracy of medication adherence forecasting. Code available: https://github.com/ab9mamun/AIMI.
GlucoLens: Explainable Postprandial Blood Glucose Prediction from Diet and Physical Activity
Mar 05, 2025Abstract:Postprandial hyperglycemia, marked by the blood glucose level exceeding the normal range after meals, is a critical indicator of progression toward type 2 diabetes in prediabetic and healthy individuals. A key metric for understanding blood glucose dynamics after eating is the postprandial area under the curve (PAUC). Predicting PAUC in advance based on a person's diet and activity level and explaining what affects postprandial blood glucose could allow an individual to adjust their lifestyle accordingly to maintain normal glucose levels. In this paper, we propose GlucoLens, an explainable machine learning approach to predict PAUC and hyperglycemia from diet, activity, and recent glucose patterns. We conducted a five-week user study with 10 full-time working individuals to develop and evaluate the computational model. Our machine learning model takes multimodal data including fasting glucose, recent glucose, recent activity, and macronutrient amounts, and provides an interpretable prediction of the postprandial glucose pattern. Our extensive analyses of the collected data revealed that the trained model achieves a normalized root mean squared error (NRMSE) of 0.123. On average, GlucoLense with a Random Forest backbone provides a 16% better result than the baseline models. Additionally, GlucoLens predicts hyperglycemia with an accuracy of 74% and recommends different options to help avoid hyperglycemia through diverse counterfactual explanations. Code available: https://github.com/ab9mamun/GlucoLens.
NutriGen: Personalized Meal Plan Generator Leveraging Large Language Models to Enhance Dietary and Nutritional Adherence
Feb 28, 2025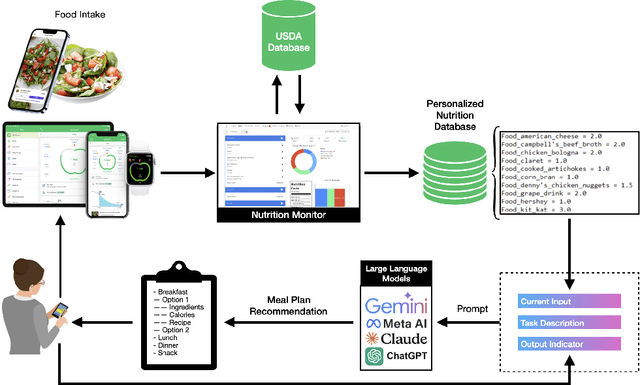
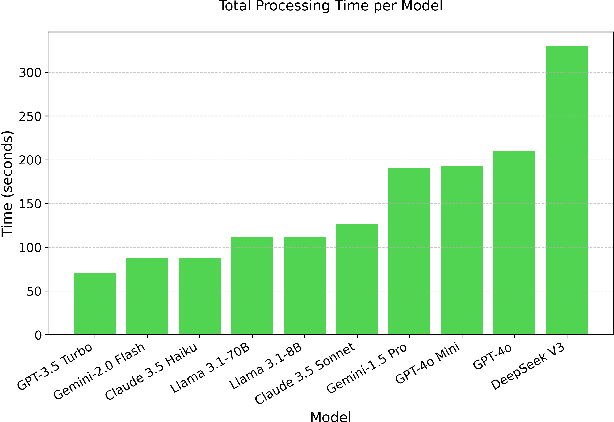
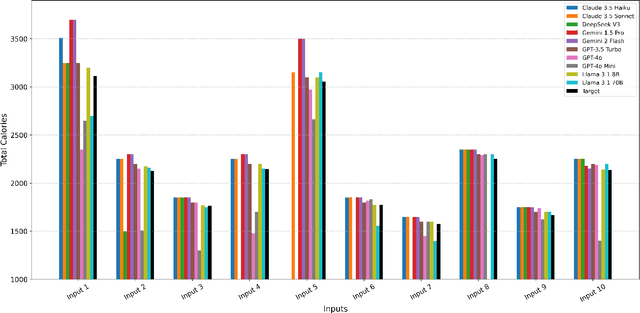
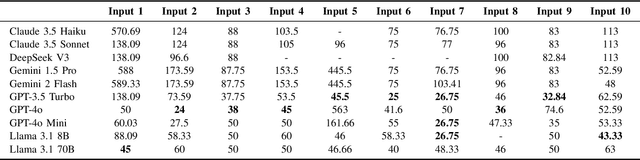
Abstract:Maintaining a balanced diet is essential for overall health, yet many individuals struggle with meal planning due to nutritional complexity, time constraints, and lack of dietary knowledge. Personalized food recommendations can help address these challenges by tailoring meal plans to individual preferences, habits, and dietary restrictions. However, existing dietary recommendation systems often lack adaptability, fail to consider real-world constraints such as food ingredient availability, and require extensive user input, making them impractical for sustainable and scalable daily use. To address these limitations, we introduce NutriGen, a framework based on large language models (LLM) designed to generate personalized meal plans that align with user-defined dietary preferences and constraints. By building a personalized nutrition database and leveraging prompt engineering, our approach enables LLMs to incorporate reliable nutritional references like the USDA nutrition database while maintaining flexibility and ease-of-use. We demonstrate that LLMs have strong potential in generating accurate and user-friendly food recommendations, addressing key limitations in existing dietary recommendation systems by providing structured, practical, and scalable meal plans. Our evaluation shows that Llama 3.1 8B and GPT-3.5 Turbo achieve the lowest percentage errors of 1.55\% and 3.68\%, respectively, producing meal plans that closely align with user-defined caloric targets while minimizing deviation and improving precision. Additionally, we compared the performance of DeepSeek V3 against several established models to evaluate its potential in personalized nutrition planning.
Type 1 Diabetes Management using GLIMMER: Glucose Level Indicator Model with Modified Error Rate
Feb 20, 2025Abstract:Managing Type 1 Diabetes (T1D) demands constant vigilance as individuals strive to regulate their blood glucose levels to avert the dangers of dysglycemia (hyperglycemia or hypoglycemia). Despite the advent of sophisticated technologies such as automated insulin delivery (AID) systems, achieving optimal glycemic control remains a formidable task. AID systems integrate continuous subcutaneous insulin infusion (CSII) and continuous glucose monitors (CGM) data, offering promise in reducing variability and increasing glucose time-in-range. However, these systems often fail to prevent dysglycemia, partly due to limitations in prediction algorithms that lack the precision to avert abnormal glucose events. This gap highlights the need for proactive behavioral adjustments. We address this need with GLIMMER, Glucose Level Indicator Model with Modified Error Rate, a machine learning approach for forecasting blood glucose levels. GLIMMER categorizes glucose values into normal and abnormal ranges and devises a novel custom loss function to prioritize accuracy in dysglycemic events where patient safety is critical. To evaluate the potential of GLIMMER for T1D management, we both use a publicly available dataset and collect new data involving 25 patients with T1D. In predicting next-hour glucose values, GLIMMER achieved a root mean square error (RMSE) of 23.97 (+/-3.77) and a mean absolute error (MAE) of 15.83 (+/-2.09) mg/dL. These results reflect a 23% improvement in RMSE and a 31% improvement in MAE compared to the best-reported error rates.
Freezing of Gait Detection Using Gramian Angular Fields and Federated Learning from Wearable Sensors
Nov 19, 2024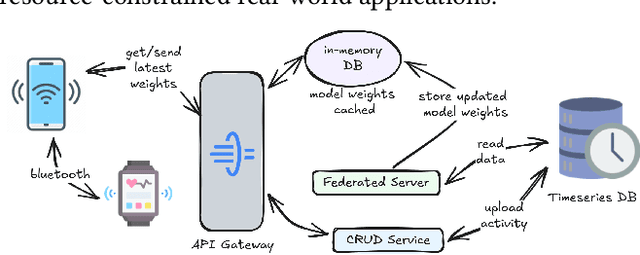

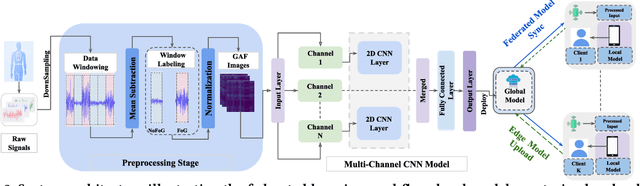

Abstract:Freezing of gait (FOG) is a debilitating symptom of Parkinson's disease (PD) that impairs mobility and safety. Traditional detection methods face challenges due to intra and inter-patient variability, and most systems are tested in controlled settings, limiting their real-world applicability. Addressing these gaps, we present FOGSense, a novel FOG detection system designed for uncontrolled, free-living conditions. It uses Gramian Angular Field (GAF) transformations and federated deep learning to capture temporal and spatial gait patterns missed by traditional methods. We evaluated our FOGSense system using a public PD dataset, 'tdcsfog'. FOGSense improves accuracy by 10.4% over a single-axis accelerometer, reduces failure points compared to multi-sensor systems, and demonstrates robustness to missing values. The federated architecture allows personalized model adaptation and efficient smartphone synchronization during off-peak hours, making it effective for long-term monitoring as symptoms evolve. Overall, FOGSense achieves a 22.2% improvement in F1-score compared to state-of-the-art methods, along with enhanced sensitivity for FOG episode detection. Code is available: https://github.com/shovito66/FOGSense.
Hybrid Attention Model Using Feature Decomposition and Knowledge Distillation for Glucose Forecasting
Nov 16, 2024Abstract:The availability of continuous glucose monitors as over-the-counter commodities have created a unique opportunity to monitor a person's blood glucose levels, forecast blood glucose trajectories and provide automated interventions to prevent devastating chronic complications that arise from poor glucose control. However, forecasting blood glucose levels is challenging because blood glucose changes consistently in response to food intake, medication intake, physical activity, sleep, and stress. It is particularly difficult to accurately predict BGL from multimodal and irregularly sampled data and over long prediction horizons. Furthermore, these forecasting models must operate in real-time on edge devices to provide in-the-moment interventions. To address these challenges, we propose GlucoNet, an AI-powered sensor system for continuously monitoring behavioral and physiological health and robust forecasting of blood glucose patterns. GlucoNet devises a feature decomposition-based transformer model that incorporates patients' behavioral and physiological data and transforms sparse and irregular patient data (e.g., diet and medication intake data) into continuous features using a mathematical model, facilitating better integration with the BGL data. Given the non-linear and non-stationary nature of BG signals, we propose a decomposition method to extract both low and high-frequency components from the BGL signals, thus providing accurate forecasting. To reduce the computational complexity, we also propose to employ knowledge distillation to compress the transformer model. GlucoNet achieves a 60% improvement in RMSE and a 21% reduction in the number of parameters, using data obtained involving 12 participants with T1-Diabetes. These results underscore GlucoNet's potential as a compact and reliable tool for real-world diabetes prevention and management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge