Asiful Arefeen
Counterfactual Modeling with Fine-Tuned LLMs for Health Intervention Design and Sensor Data Augmentation
Jan 21, 2026Abstract:Counterfactual explanations (CFEs) provide human-centric interpretability by identifying the minimal, actionable changes required to alter a machine learning model's prediction. Therefore, CFs can be used as (i) interventions for abnormality prevention and (ii) augmented data for training robust models. We conduct a comprehensive evaluation of CF generation using large language models (LLMs), including GPT-4 (zero-shot and few-shot) and two open-source models-BioMistral-7B and LLaMA-3.1-8B, in both pretrained and fine-tuned configurations. Using the multimodal AI-READI clinical dataset, we assess CFs across three dimensions: intervention quality, feature diversity, and augmentation effectiveness. Fine-tuned LLMs, particularly LLaMA-3.1-8B, produce CFs with high plausibility (up to 99%), strong validity (up to 0.99), and realistic, behaviorally modifiable feature adjustments. When used for data augmentation under controlled label-scarcity settings, LLM-generated CFs substantially restore classifier performance, yielding an average 20% F1 recovery across three scarcity scenarios. Compared with optimization-based baselines such as DiCE, CFNOW, and NICE, LLMs offer a flexible, model-agnostic approach that generates more clinically actionable and semantically coherent counterfactuals. Overall, this work demonstrates the promise of LLM-driven counterfactuals for both interpretable intervention design and data-efficient model training in sensor-based digital health. Impact: SenseCF fine-tunes an LLM to generate valid, representative counterfactual explanations and supplement minority class in an imbalanced dataset for improving model training and boosting model robustness and predictive performance
GlyTwin: Digital Twin for Glucose Control in Type 1 Diabetes Through Optimal Behavioral Modifications Using Patient-Centric Counterfactuals
Apr 14, 2025Abstract:Frequent and long-term exposure to hyperglycemia (i.e., high blood glucose) increases the risk of chronic complications such as neuropathy, nephropathy, and cardiovascular disease. Current technologies like continuous subcutaneous insulin infusion (CSII) and continuous glucose monitoring (CGM) primarily model specific aspects of glycemic control-like hypoglycemia prediction or insulin delivery. Similarly, most digital twin approaches in diabetes management simulate only physiological processes. These systems lack the ability to offer alternative treatment scenarios that support proactive behavioral interventions. To address this, we propose GlyTwin, a novel digital twin framework that uses counterfactual explanations to simulate optimal treatments for glucose regulation. Our approach helps patients and caregivers modify behaviors like carbohydrate intake and insulin dosing to avoid abnormal glucose events. GlyTwin generates behavioral treatment suggestions that proactively prevent hyperglycemia by recommending small adjustments to daily choices, reducing both frequency and duration of these events. Additionally, it incorporates stakeholder preferences into the intervention design, making recommendations patient-centric and tailored. We evaluate GlyTwin on AZT1D, a newly constructed dataset with longitudinal data from 21 type 1 diabetes (T1D) patients on automated insulin delivery systems over 26 days. Results show GlyTwin outperforms state-of-the-art counterfactual methods, generating 76.6% valid and 86% effective interventions. These findings demonstrate the promise of counterfactual-driven digital twins in delivering personalized healthcare.
GlucoLens: Explainable Postprandial Blood Glucose Prediction from Diet and Physical Activity
Mar 05, 2025Abstract:Postprandial hyperglycemia, marked by the blood glucose level exceeding the normal range after meals, is a critical indicator of progression toward type 2 diabetes in prediabetic and healthy individuals. A key metric for understanding blood glucose dynamics after eating is the postprandial area under the curve (PAUC). Predicting PAUC in advance based on a person's diet and activity level and explaining what affects postprandial blood glucose could allow an individual to adjust their lifestyle accordingly to maintain normal glucose levels. In this paper, we propose GlucoLens, an explainable machine learning approach to predict PAUC and hyperglycemia from diet, activity, and recent glucose patterns. We conducted a five-week user study with 10 full-time working individuals to develop and evaluate the computational model. Our machine learning model takes multimodal data including fasting glucose, recent glucose, recent activity, and macronutrient amounts, and provides an interpretable prediction of the postprandial glucose pattern. Our extensive analyses of the collected data revealed that the trained model achieves a normalized root mean squared error (NRMSE) of 0.123. On average, GlucoLense with a Random Forest backbone provides a 16% better result than the baseline models. Additionally, GlucoLens predicts hyperglycemia with an accuracy of 74% and recommends different options to help avoid hyperglycemia through diverse counterfactual explanations. Code available: https://github.com/ab9mamun/GlucoLens.
NutriGen: Personalized Meal Plan Generator Leveraging Large Language Models to Enhance Dietary and Nutritional Adherence
Feb 28, 2025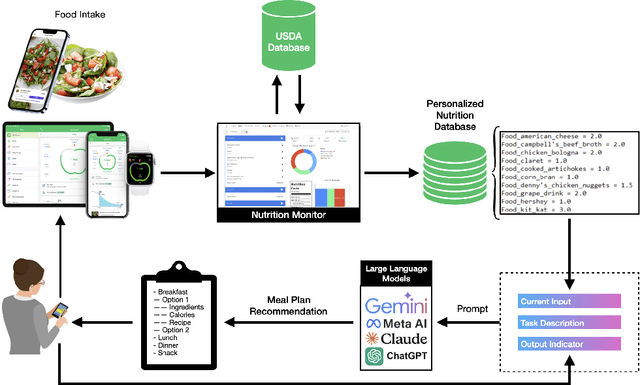
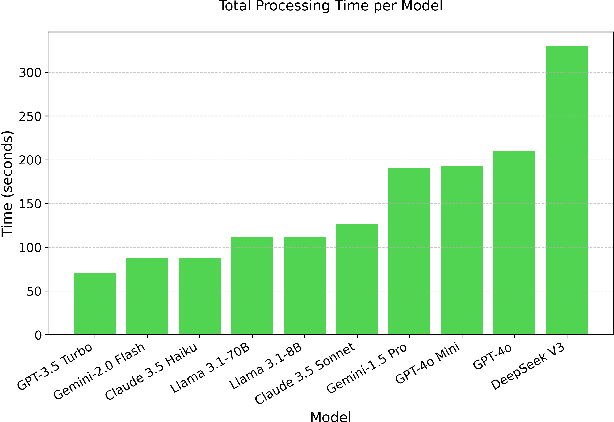
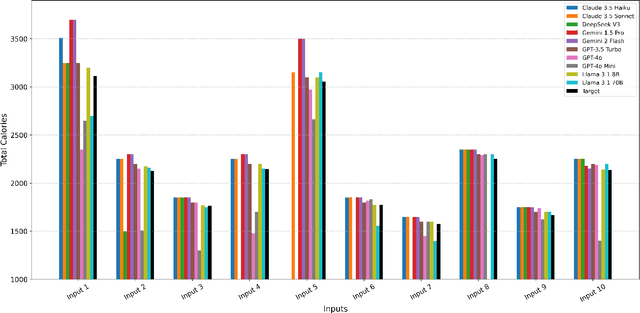
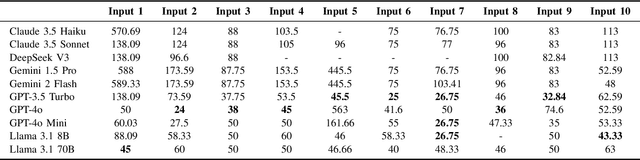
Abstract:Maintaining a balanced diet is essential for overall health, yet many individuals struggle with meal planning due to nutritional complexity, time constraints, and lack of dietary knowledge. Personalized food recommendations can help address these challenges by tailoring meal plans to individual preferences, habits, and dietary restrictions. However, existing dietary recommendation systems often lack adaptability, fail to consider real-world constraints such as food ingredient availability, and require extensive user input, making them impractical for sustainable and scalable daily use. To address these limitations, we introduce NutriGen, a framework based on large language models (LLM) designed to generate personalized meal plans that align with user-defined dietary preferences and constraints. By building a personalized nutrition database and leveraging prompt engineering, our approach enables LLMs to incorporate reliable nutritional references like the USDA nutrition database while maintaining flexibility and ease-of-use. We demonstrate that LLMs have strong potential in generating accurate and user-friendly food recommendations, addressing key limitations in existing dietary recommendation systems by providing structured, practical, and scalable meal plans. Our evaluation shows that Llama 3.1 8B and GPT-3.5 Turbo achieve the lowest percentage errors of 1.55\% and 3.68\%, respectively, producing meal plans that closely align with user-defined caloric targets while minimizing deviation and improving precision. Additionally, we compared the performance of DeepSeek V3 against several established models to evaluate its potential in personalized nutrition planning.
Type 1 Diabetes Management using GLIMMER: Glucose Level Indicator Model with Modified Error Rate
Feb 20, 2025Abstract:Managing Type 1 Diabetes (T1D) demands constant vigilance as individuals strive to regulate their blood glucose levels to avert the dangers of dysglycemia (hyperglycemia or hypoglycemia). Despite the advent of sophisticated technologies such as automated insulin delivery (AID) systems, achieving optimal glycemic control remains a formidable task. AID systems integrate continuous subcutaneous insulin infusion (CSII) and continuous glucose monitors (CGM) data, offering promise in reducing variability and increasing glucose time-in-range. However, these systems often fail to prevent dysglycemia, partly due to limitations in prediction algorithms that lack the precision to avert abnormal glucose events. This gap highlights the need for proactive behavioral adjustments. We address this need with GLIMMER, Glucose Level Indicator Model with Modified Error Rate, a machine learning approach for forecasting blood glucose levels. GLIMMER categorizes glucose values into normal and abnormal ranges and devises a novel custom loss function to prioritize accuracy in dysglycemic events where patient safety is critical. To evaluate the potential of GLIMMER for T1D management, we both use a publicly available dataset and collect new data involving 25 patients with T1D. In predicting next-hour glucose values, GLIMMER achieved a root mean square error (RMSE) of 23.97 (+/-3.77) and a mean absolute error (MAE) of 15.83 (+/-2.09) mg/dL. These results reflect a 23% improvement in RMSE and a 31% improvement in MAE compared to the best-reported error rates.
Designing User-Centric Behavioral Interventions to Prevent Dysglycemia with Novel Counterfactual Explanations
Oct 02, 2023Abstract:Maintaining normal blood glucose levels through lifestyle behaviors is central to maintaining health and preventing disease. Frequent exposure to dysglycemia (i.e., abnormal glucose events such as hyperlycemia and hypoglycemia) leads to chronic complications including diabetes, kidney disease and need for dialysis, myocardial infarction, stroke, amputation, and death. Therefore, a tool capable of predicting dysglycemia and offering users actionable feedback about how to make changes in their diet, exercise, and medication to prevent abnormal glycemic events could have significant societal impacts. Counterfactual explanations can provide insights into why a model made a particular prediction by generating hypothetical instances that are similar to the original input but lead to a different prediction outcome. Therefore, counterfactuals can be viewed as a means to design AI-driven health interventions to prevent adverse health outcomes such as dysglycemia. In this paper, we design GlyCoach, a framework for generating counterfactual explanations for glucose control. Leveraging insights from adversarial learning, GlyCoach characterizes the decision boundary for high-dimensional health data and performs a grid search to generate actionable interventions. GlyCoach is unique in integrating prior knowledge about user preferences of plausible explanations into the process of counterfactual generation. We evaluate GlyCoach extensively using two real-world datasets and external simulators from prior studies that predict glucose response. GlyCoach achieves 87\% sensitivity in the simulation-aided validation, surpassing the state-of-the-art techniques for generating counterfactual explanations by at least $10\%$. Besides, counterfactuals from GlyCoach exhibit a $32\%$ improved normalized distance compared to previous research.
Inter-Beat Interval Estimation with Tiramisu Model: A Novel Approach with Reduced Error
Jul 01, 2021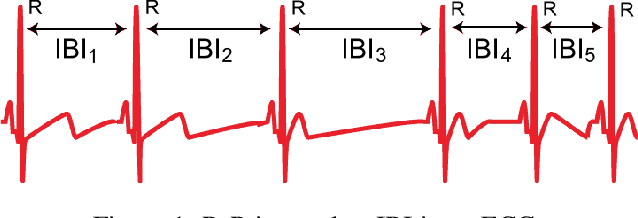
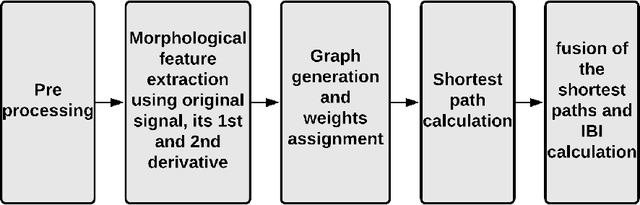
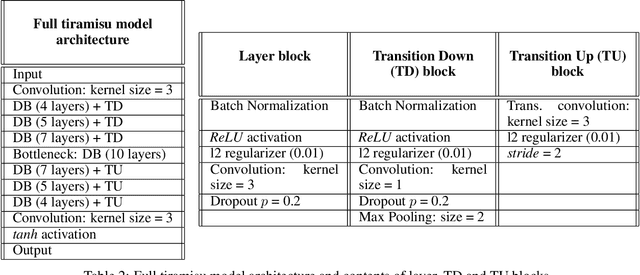
Abstract:Inter-beat interval (IBI) measurement enables estimation of heart-rate variability (HRV) which, in turns, can provide early indication of potential cardiovascular diseases. However, extracting IBIs from noisy signals is challenging since the morphology of the signal is distorted in the presence of the noise. Electrocardiogram (ECG) of a person in heavy motion is highly corrupted with noise, known as motion-artifact, and IBI extracted from it is inaccurate. As a part of remote health monitoring and wearable system development, denoising ECG signals and estimating IBIs correctly from them have become an emerging topic among signal-processing researchers. Apart from conventional methods, deep-learning techniques have been successfully used in signal denoising recently, and diagnosis process has become easier, leading to accuracy levels that were previously unachievable. We propose a deep-learning approach leveraging tiramisu autoencoder model to suppress motion-artifact noise and make the R-peaks of the ECG signal prominent even in the presence of high-intensity motion. After denoising, IBIs are estimated more accurately expediting diagnosis tasks. Results illustrate that our method enables IBI estimation from noisy ECG signals with SNR up to -30dB with average root mean square error (RMSE) of 13 milliseconds for estimated IBIs. At this noise level, our error percentage remains below 8% and outperforms other state of the art techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge