Mario Sänger
Knowledge-augmented Pre-trained Language Models for Biomedical Relation Extraction
May 01, 2025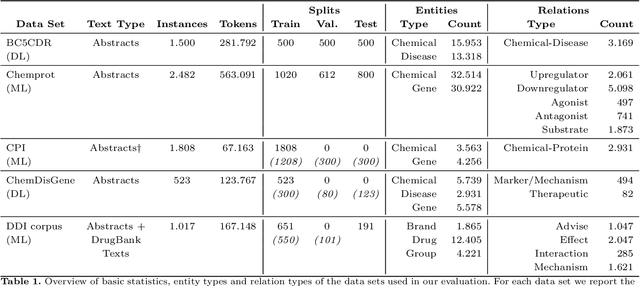
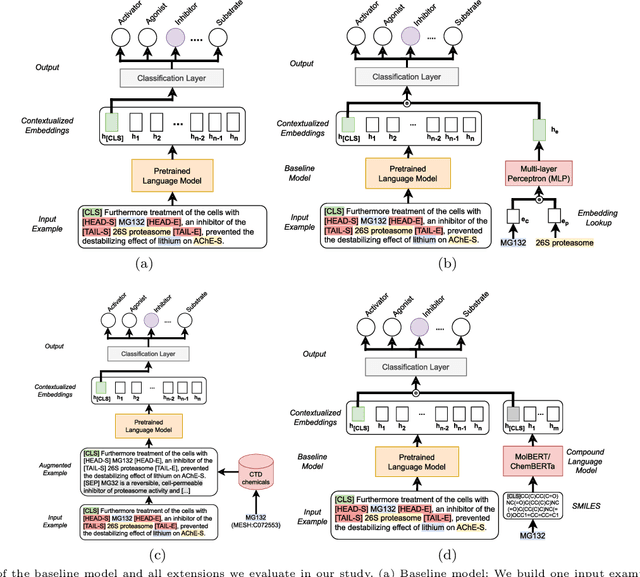
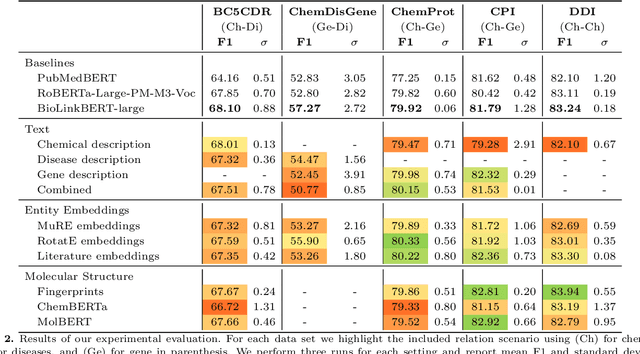
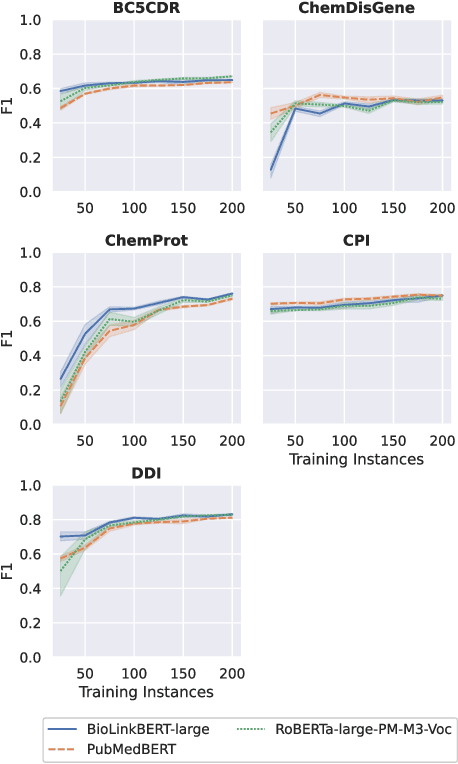
Abstract:Automatic relationship extraction (RE) from biomedical literature is critical for managing the vast amount of scientific knowledge produced each year. In recent years, utilizing pre-trained language models (PLMs) has become the prevalent approach in RE. Several studies report improved performance when incorporating additional context information while fine-tuning PLMs for RE. However, variations in the PLMs applied, the databases used for augmentation, hyper-parameter optimization, and evaluation methods complicate direct comparisons between studies and raise questions about the generalizability of these findings. Our study addresses this research gap by evaluating PLMs enhanced with contextual information on five datasets spanning four relation scenarios within a consistent evaluation framework. We evaluate three baseline PLMs and first conduct extensive hyperparameter optimization. After selecting the top-performing model, we enhance it with additional data, including textual entity descriptions, relational information from knowledge graphs, and molecular structure encodings. Our findings illustrate the importance of i) the choice of the underlying language model and ii) a comprehensive hyperparameter optimization for achieving strong extraction performance. Although inclusion of context information yield only minor overall improvements, an ablation study reveals substantial benefits for smaller PLMs when such external data was included during fine-tuning.
HunFlair2 in a cross-corpus evaluation of biomedical named entity recognition and normalization tools
Feb 20, 2024Abstract:With the exponential growth of the life science literature, biomedical text mining (BTM) has become an essential technology for accelerating the extraction of insights from publications. Identifying named entities (e.g., diseases, drugs, or genes) in texts and their linkage to reference knowledge bases are crucial steps in BTM pipelines to enable information aggregation from different documents. However, tools for these two steps are rarely applied in the same context in which they were developed. Instead, they are applied in the wild, i.e., on application-dependent text collections different from those used for the tools' training, varying, e.g., in focus, genre, style, and text type. This raises the question of whether the reported performance of BTM tools can be trusted for downstream applications. Here, we report on the results of a carefully designed cross-corpus benchmark for named entity extraction, where tools were applied systematically to corpora not used during their training. Based on a survey of 28 published systems, we selected five for an in-depth analysis on three publicly available corpora encompassing four different entity types. Comparison between tools results in a mixed picture and shows that, in a cross-corpus setting, the performance is significantly lower than the one reported in an in-corpus setting. HunFlair2 showed the best performance on average, being closely followed by PubTator. Our results indicate that users of BTM tools should expect diminishing performances when applying them in the wild compared to original publications and show that further research is necessary to make BTM tools more robust.
Large Language Models to the Rescue: Reducing the Complexity in Scientific Workflow Development Using ChatGPT
Nov 06, 2023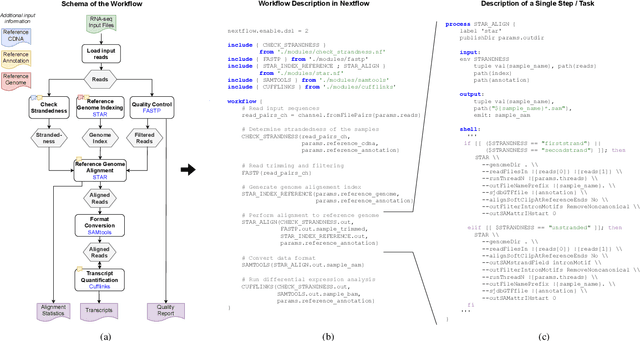
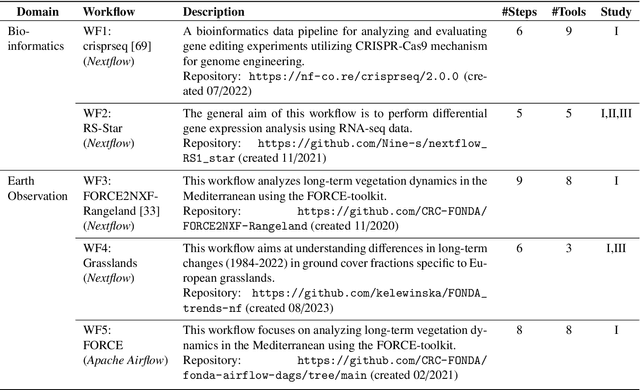

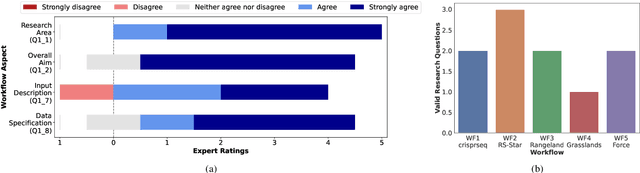
Abstract:Scientific workflow systems are increasingly popular for expressing and executing complex data analysis pipelines over large datasets, as they offer reproducibility, dependability, and scalability of analyses by automatic parallelization on large compute clusters. However, implementing workflows is difficult due to the involvement of many black-box tools and the deep infrastructure stack necessary for their execution. Simultaneously, user-supporting tools are rare, and the number of available examples is much lower than in classical programming languages. To address these challenges, we investigate the efficiency of Large Language Models (LLMs), specifically ChatGPT, to support users when dealing with scientific workflows. We performed three user studies in two scientific domains to evaluate ChatGPT for comprehending, adapting, and extending workflows. Our results indicate that LLMs efficiently interpret workflows but achieve lower performance for exchanging components or purposeful workflow extensions. We characterize their limitations in these challenging scenarios and suggest future research directions.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022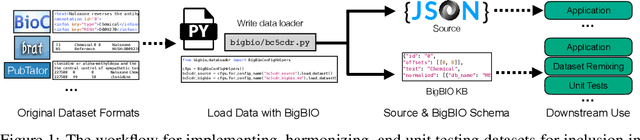
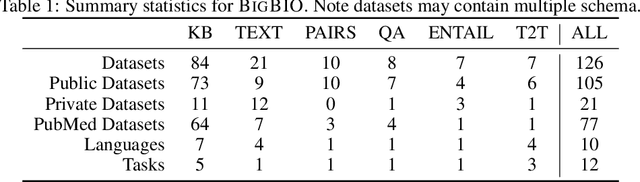
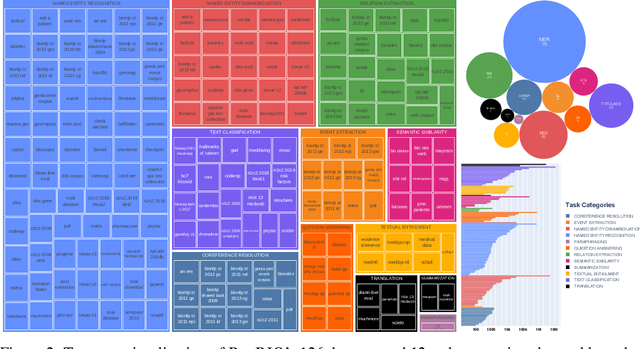
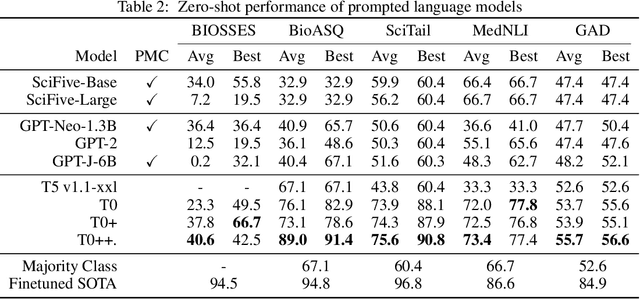
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
HunFlair: An Easy-to-Use Tool for State-of-the-Art Biomedical Named Entity Recognition
Aug 18, 2020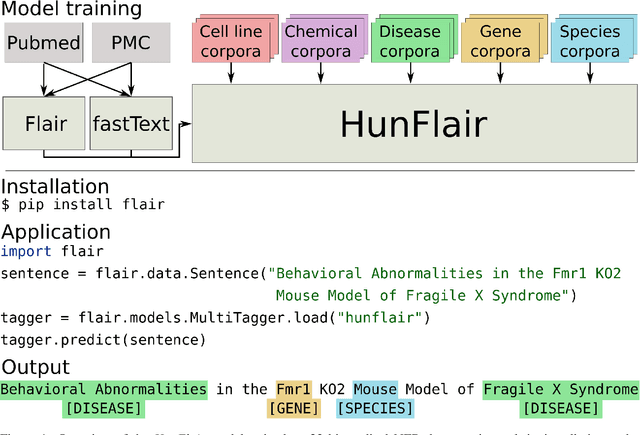
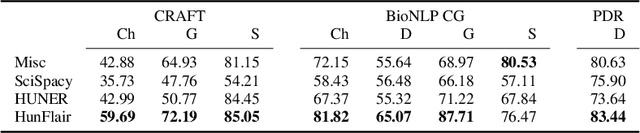
Abstract:Summary: Named Entity Recognition (NER) is an important step in biomedical information extraction pipelines. Tools for NER should be easy to use, cover multiple entity types, highly accurate, and robust towards variations in text genre and style. To this end, we propose HunFlair, an NER tagger covering multiple entity types integrated into the widely used NLP framework Flair. HunFlair outperforms other state-of-the-art standalone NER tools with an average gain of 7.26 pp over the next best tool, can be installed with a single command and is applied with only four lines of code. Availability: HunFlair is freely available through the Flair framework under an MIT license: https://github.com/flairNLP/flair and is compatible with all major operating systems. Contact:{weberple,saengema,alan.akbik}@informatik.hu-berlin.de
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge