Li Xie
Step-DeepResearch Technical Report
Dec 24, 2025Abstract:As LLMs shift toward autonomous agents, Deep Research has emerged as a pivotal metric. However, existing academic benchmarks like BrowseComp often fail to meet real-world demands for open-ended research, which requires robust skills in intent recognition, long-horizon decision-making, and cross-source verification. To address this, we introduce Step-DeepResearch, a cost-effective, end-to-end agent. We propose a Data Synthesis Strategy Based on Atomic Capabilities to reinforce planning and report writing, combined with a progressive training path from agentic mid-training to SFT and RL. Enhanced by a Checklist-style Judger, this approach significantly improves robustness. Furthermore, to bridge the evaluation gap in the Chinese domain, we establish ADR-Bench for realistic deep research scenarios. Experimental results show that Step-DeepResearch (32B) scores 61.4% on Scale AI Research Rubrics. On ADR-Bench, it significantly outperforms comparable models and rivals SOTA closed-source models like OpenAI and Gemini DeepResearch. These findings prove that refined training enables medium-sized models to achieve expert-level capabilities at industry-leading cost-efficiency.
Co-Sight: Enhancing LLM-Based Agents via Conflict-Aware Meta-Verification and Trustworthy Reasoning with Structured Facts
Oct 24, 2025Abstract:Long-horizon reasoning in LLM-based agents often fails not from generative weakness but from insufficient verification of intermediate reasoning. Co-Sight addresses this challenge by turning reasoning into a falsifiable and auditable process through two complementary mechanisms: Conflict-Aware Meta-Verification (CAMV) and Trustworthy Reasoning with Structured Facts (TRSF). CAMV reformulates verification as conflict identification and targeted falsification, allocating computation only to disagreement hotspots among expert agents rather than to full reasoning chains. This bounds verification cost to the number of inconsistencies and improves efficiency and reliability. TRSF continuously organizes, validates, and synchronizes evidence across agents through a structured facts module. By maintaining verified, traceable, and auditable knowledge, it ensures that all reasoning is grounded in consistent, source-verified information and supports transparent verification throughout the reasoning process. Together, TRSF and CAMV form a closed verification loop, where TRSF supplies structured facts and CAMV selectively falsifies or reinforces them, yielding transparent and trustworthy reasoning. Empirically, Co-Sight achieves state-of-the-art accuracy on GAIA (84.4%) and Humanity's Last Exam (35.5%), and strong results on Chinese-SimpleQA (93.8%). Ablation studies confirm that the synergy between structured factual grounding and conflict-aware verification drives these improvements. Co-Sight thus offers a scalable paradigm for reliable long-horizon reasoning in LLM-based agents. Code is available at https://github.com/ZTE-AICloud/Co-Sight/tree/cosight2.0_benchmarks.
Data Quality Enhancement on the Basis of Diversity with Large Language Models for Text Classification: Uncovered, Difficult, and Noisy
Dec 10, 2024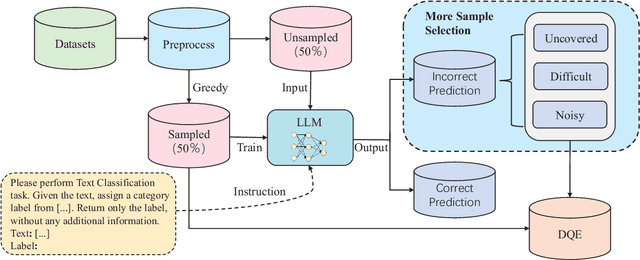
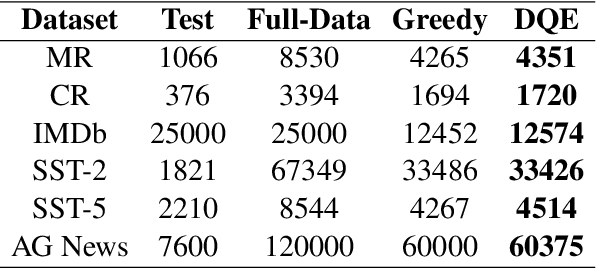
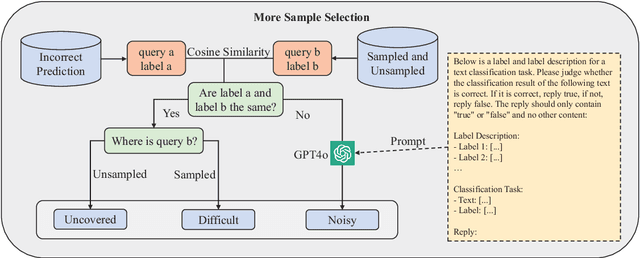
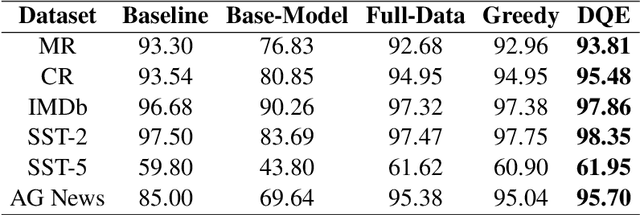
Abstract:In recent years, the use of large language models (LLMs) for text classification has attracted widespread attention. Despite this, the classification accuracy of LLMs has not yet universally surpassed that of smaller models. LLMs can enhance their performance in text classification through fine-tuning. However, existing data quality research based on LLMs is challenging to apply directly to solve text classification problems. To further improve the performance of LLMs in classification tasks, this paper proposes a data quality enhancement (DQE) method for text classification based on LLMs. This method starts by using a greedy algorithm to select data, dividing the dataset into sampled and unsampled subsets, and then performing fine-tuning of the LLMs using the sampled data. Subsequently, this model is used to predict the outcomes for the unsampled data, categorizing incorrectly predicted data into uncovered, difficult, and noisy data. Experimental results demonstrate that our method effectively enhances the performance of LLMs in text classification tasks and significantly improves training efficiency, saving nearly half of the training time. Our method has achieved state-of-the-art performance in several open-source classification tasks.
Gaussian Rate-Distortion-Perception Coding and Entropy-Constrained Scalar Quantization
Sep 04, 2024



Abstract:This paper investigates the best known bounds on the quadratic Gaussian distortion-rate-perception function with limited common randomness for the Kullback-Leibler divergence-based perception measure, as well as their counterparts for the squared Wasserstein-2 distance-based perception measure, recently established by Xie et al. These bounds are shown to be nondegenerate in the sense that they cannot be deduced from each other via a refined version of Talagrand's transportation inequality. On the other hand, an improved lower bound is established when the perception measure is given by the squared Wasserstein-2 distance. In addition, it is revealed by exploiting the connection between rate-distortion-perception coding and entropy-constrained scalar quantization that all the aforementioned bounds are generally not tight in the weak perception constraint regime.
Output-Constrained Lossy Source Coding With Application to Rate-Distortion-Perception Theory
Mar 21, 2024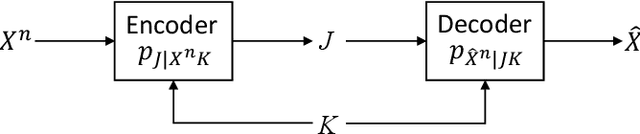
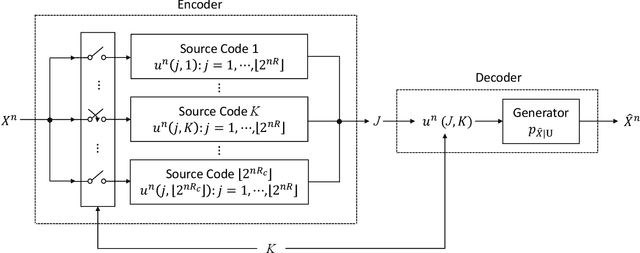
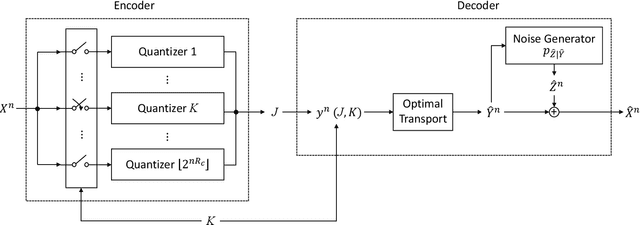
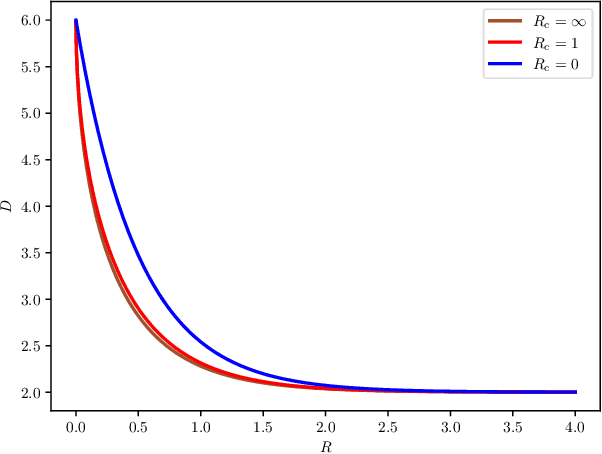
Abstract:The distortion-rate function of output-constrained lossy source coding with limited common randomness is analyzed for the special case of squared error distortion measure. An explicit expression is obtained when both source and reconstruction distributions are Gaussian. This further leads to a partial characterization of the information-theoretic limit of quadratic Gaussian rate-distortion-perception coding with the perception measure given by Kullback-Leibler divergence or squared quadratic Wasserstein distance.
Learning Universal and Robust 3D Molecular Representations with Graph Convolutional Networks
Jul 24, 2023Abstract:To learn accurate representations of molecules, it is essential to consider both chemical and geometric features. To encode geometric information, many descriptors have been proposed in constrained circumstances for specific types of molecules and do not have the properties to be ``robust": 1. Invariant to rotations and translations; 2. Injective when embedding molecular structures. In this work, we propose a universal and robust Directional Node Pair (DNP) descriptor based on the graph representations of 3D molecules. Our DNP descriptor is robust compared to previous ones and can be applied to multiple molecular types. To combine the DNP descriptor and chemical features in molecules, we construct the Robust Molecular Graph Convolutional Network (RoM-GCN) which is capable to take both node and edge features into consideration when generating molecule representations. We evaluate our model on protein and small molecule datasets. Our results validate the superiority of the DNP descriptor in incorporating 3D geometric information of molecules. RoM-GCN outperforms all compared baselines.
Exploration of Dark Chemical Genomics Space via Portal Learning: Applied to Targeting the Undruggable Genome and COVID-19 Anti-Infective Polypharmacology
Nov 23, 2021
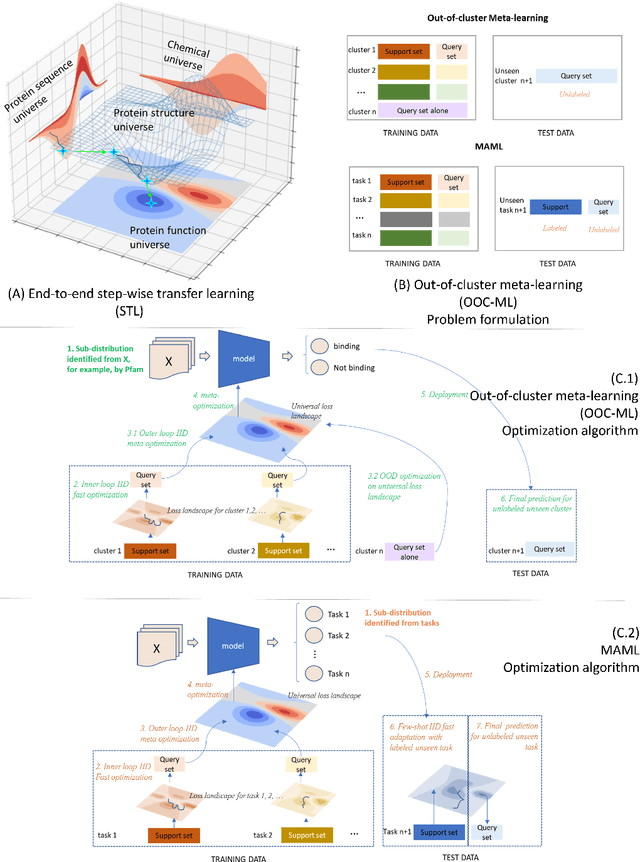
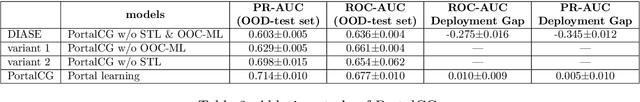
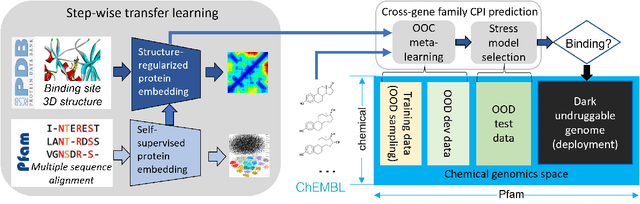
Abstract:Advances in biomedicine are largely fueled by exploring uncharted territories of human biology. Machine learning can both enable and accelerate discovery, but faces a fundamental hurdle when applied to unseen data with distributions that differ from previously observed ones -- a common dilemma in scientific inquiry. We have developed a new deep learning framework, called {\textit{Portal Learning}}, to explore dark chemical and biological space. Three key, novel components of our approach include: (i) end-to-end, step-wise transfer learning, in recognition of biology's sequence-structure-function paradigm, (ii) out-of-cluster meta-learning, and (iii) stress model selection. Portal Learning provides a practical solution to the out-of-distribution (OOD) problem in statistical machine learning. Here, we have implemented Portal Learning to predict chemical-protein interactions on a genome-wide scale. Systematic studies demonstrate that Portal Learning can effectively assign ligands to unexplored gene families (unknown functions), versus existing state-of-the-art methods, thereby allowing us to target previously "undruggable" proteins and design novel polypharmacological agents for disrupting interactions between SARS-CoV-2 and human proteins. Portal Learning is general-purpose and can be further applied to other areas of scientific inquiry.
A multi-center prospective evaluation of THEIA to detect diabetic retinopathy (DR) and diabetic macular edema (DME) in the New Zealand screening program
Jun 23, 2021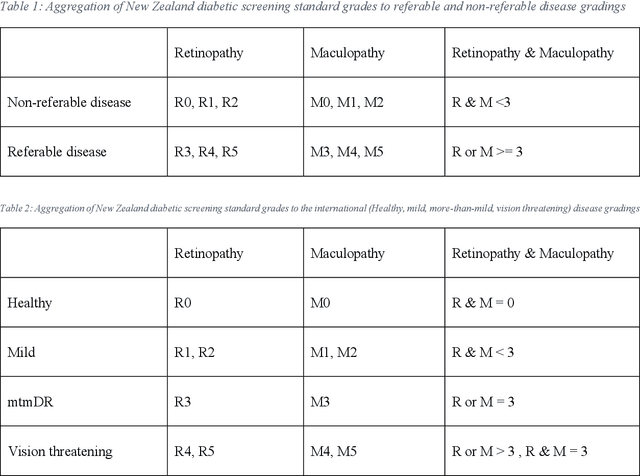
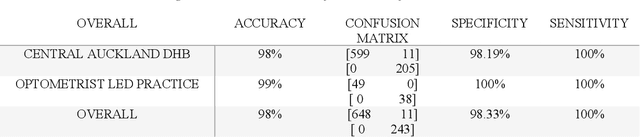
Abstract:Purpose: to assess the efficacy of THEIA, an artificial intelligence for screening diabetic retinopathy in a multi-center prospective study. To validate the potential application of THEIA as clinical decision making assistant in a national screening program. Methods: 902 patients were recruited from either an urban large eye hospital, or a semi-rural optometrist led screening provider, as they were attending their appointment as part of New Zealand Diabetic Screening programme. These clinics used a variety of retinal cameras and a range of operators. The de-identified images were then graded independently by three senior retinal specialists, and final results were aggregated using New Zealand grading scheme, which is then converted to referable\non-referable and Healthy\mild\more than mild\vision threatening categories. Results: compared to ground truth, THEIA achieved 100% sensitivity and [95.35%-97.44%] specificity, and negative predictive value of 100%. THEIA also did not miss any patients with more than mild or vision threatening disease. The level of agreement between the clinicians and the aggregated results was (k value: 0.9881, 0.9557, and 0.9175), and the level of agreement between THEIA and the aggregated labels was (k value: 0.9515). Conclusion: Our multi-centre prospective trial showed that THEIA does not miss referable disease when screening for diabetic retinopathy and maculopathy. It also has a very high level of granularity in reporting the disease level. Since THEIA is being tested on a variety of cameras, operating in a range of clinics (rural\urban, ophthalmologist-led\optometrist-led), we believe that it will be a suitable addition to a public diabetic screening program.
Real-world plant species identification based on deep convolutional neural networks and visual attention
Jul 06, 2018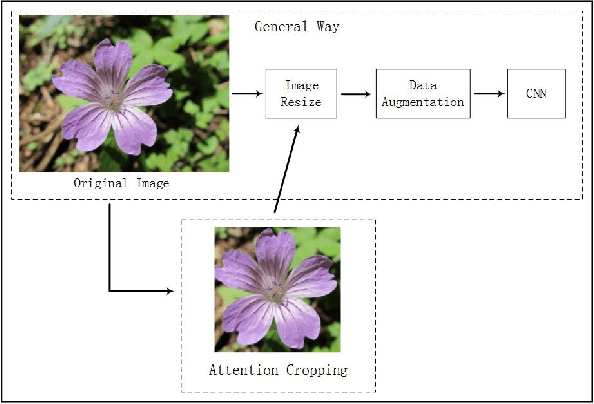
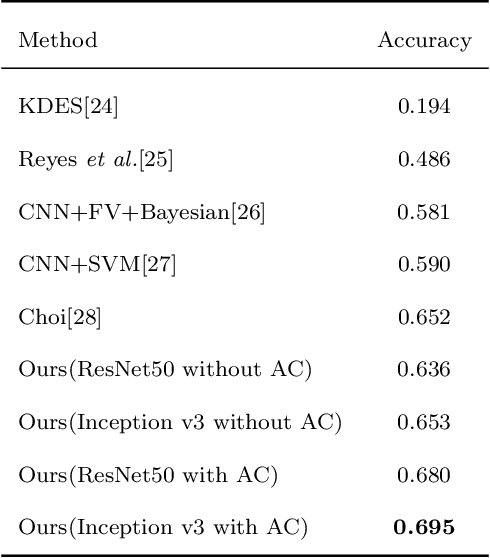
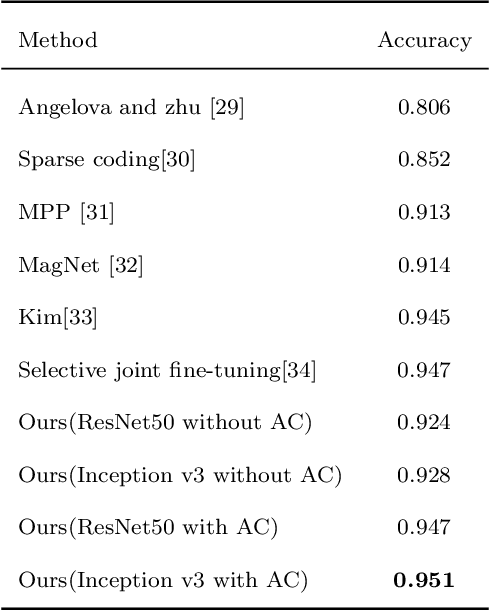
Abstract:This paper investigates the issue of real-world identification to fulfill better species protection. We focus on plant species identification as it is a classic and hot issue. In tradition plant species identification the samples are scanned specimen and the background is simple. However, real-world species recognition is more challenging. We first systematically investigate what is realistic species recognition and the difference from tradition plant species recognition. To deal with the challenging task, an interdisciplinary collaboration is presented based on the latest advances in computer science and technology. We propose a novel framework and an effective data augmentation method for deep learning in this paper. We first crop the image in terms with visual attention before general recognition. Besides, we apply it as a data augmentation method. We call the novel data augmentation approach attention cropping (AC). Deep convolutional neural networks are trained to predict species from a large amount of data. Extensive experiments on traditional dataset and specific dataset for real-world recognition are conducted to evaluate the performance of our approach. Experiments first demonstrate that our approach achieves state-of-the-art results on different types of datasets. Besides, we also evaluate the performance of data augmentation method AC. Results show that AC provides superior performance. Compared with the precision of methods without AC, the results with AC achieve substantial improvement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge