Kiran Kokilepersaud
Countering Multi-modal Representation Collapse through Rank-targeted Fusion
Nov 09, 2025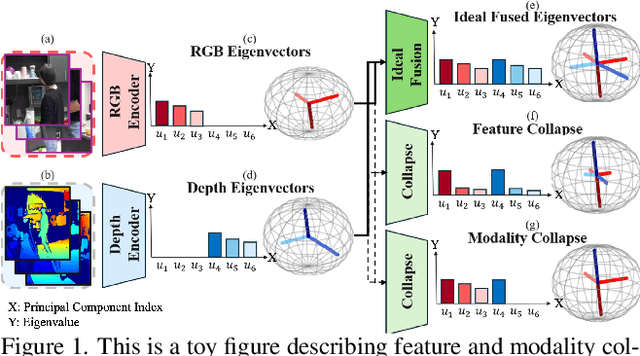
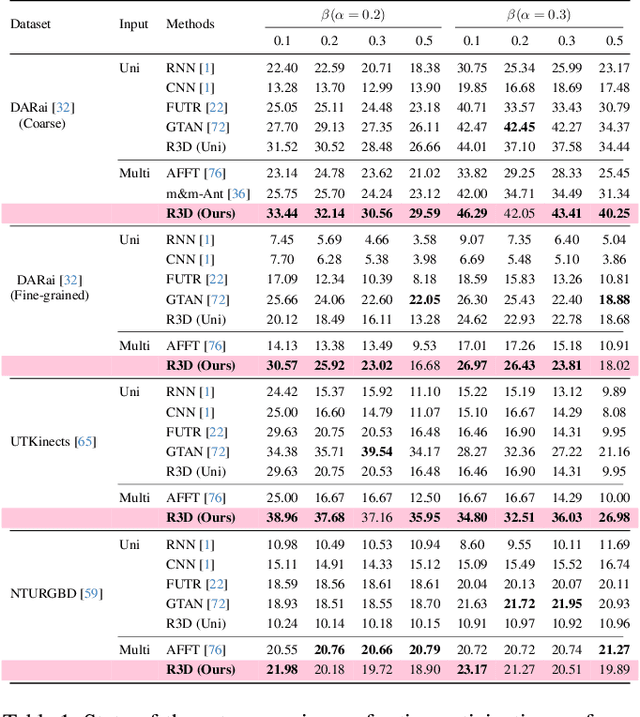
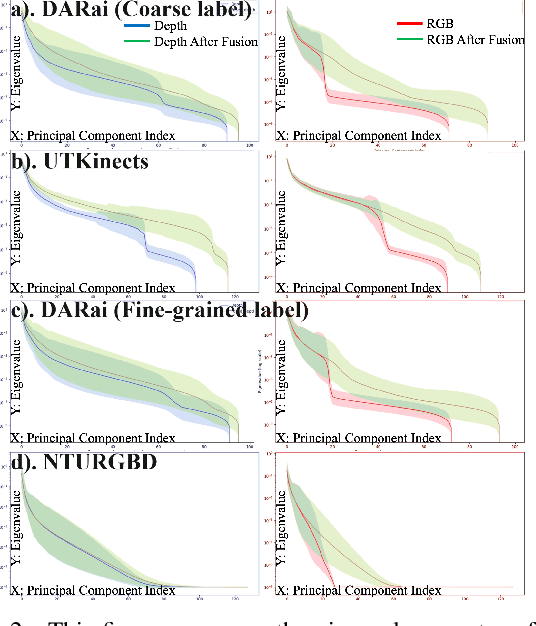
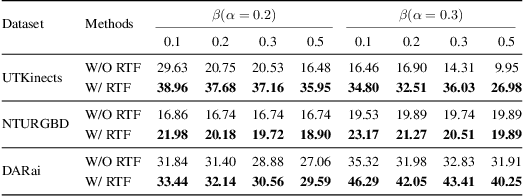
Abstract:Multi-modal fusion methods often suffer from two types of representation collapse: feature collapse where individual dimensions lose their discriminative power (as measured by eigenspectra), and modality collapse where one dominant modality overwhelms the other. Applications like human action anticipation that require fusing multifarious sensor data are hindered by both feature and modality collapse. However, existing methods attempt to counter feature collapse and modality collapse separately. This is because there is no unifying framework that efficiently addresses feature and modality collapse in conjunction. In this paper, we posit the utility of effective rank as an informative measure that can be utilized to quantify and counter both the representation collapses. We propose \textit{Rank-enhancing Token Fuser}, a theoretically grounded fusion framework that selectively blends less informative features from one modality with complementary features from another modality. We show that our method increases the effective rank of the fused representation. To address modality collapse, we evaluate modality combinations that mutually increase each others' effective rank. We show that depth maintains representational balance when fused with RGB, avoiding modality collapse. We validate our method on action anticipation, where we present \texttt{R3D}, a depth-informed fusion framework. Extensive experiments on NTURGBD, UTKinect, and DARai demonstrate that our approach significantly outperforms prior state-of-the-art methods by up to 3.74\%. Our code is available at: \href{https://github.com/olivesgatech/R3D}{https://github.com/olivesgatech/R3D}.
AdaDim: Dimensionality Adaptation for SSL Representational Dynamics
May 18, 2025Abstract:A key factor in effective Self-Supervised learning (SSL) is preventing dimensional collapse, which is where higher-dimensional representation spaces span a lower-dimensional subspace. Therefore, SSL optimization strategies involve guiding a model to produce representations ($R$) with a higher dimensionality. Dimensionality is either optimized through a dimension-contrastive approach that encourages feature decorrelation or through a sample-contrastive method that promotes a uniform spread of sample representations. Both families of SSL algorithms also utilize a projection head that maps $R$ into a lower-dimensional embedding space $Z$. Recent work has characterized the projection head as a filter of irrelevant features from the SSL objective by reducing mutual information, $I(R;Z)$. Therefore, the current literature's view is that a good SSL representation space should have a high $H(R)$ and a low $I(R;Z)$. However, this view of the problem is lacking in terms of an understanding of the underlying training dynamics that influences both terms, as well as how the values of $H(R)$ and $I(R;Z)$ arrived at the end of training reflect the downstream performance of an SSL model. We address both gaps in the literature by demonstrating that increases in $H(R)$ due to feature decorrelation at the start of training lead to a higher $I(R;Z)$, while increases in $H(R)$ due to samples distributing uniformly in a high-dimensional space at the end of training cause $I(R;Z)$ to plateau or decrease. Furthermore, our analysis shows that the best performing SSL models do not have the highest $H(R)$ nor the lowest $I(R;Z)$, but arrive at an optimal intermediate point for both. We develop a method called AdaDim to exploit these observed training dynamics by adaptively weighting between losses based on feature decorrelation and uniform sample spread.
HEX: Hierarchical Emergence Exploitation in Self-Supervised Algorithms
Oct 30, 2024
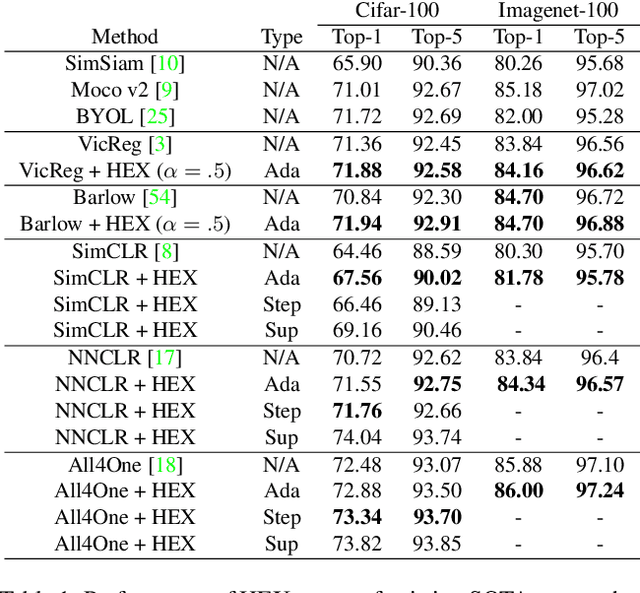

Abstract:In this paper, we propose an algorithm that can be used on top of a wide variety of self-supervised (SSL) approaches to take advantage of hierarchical structures that emerge during training. SSL approaches typically work through some invariance term to ensure consistency between similar samples and a regularization term to prevent global dimensional collapse. Dimensional collapse refers to data representations spanning a lower-dimensional subspace. Recent work has demonstrated that the representation space of these algorithms gradually reflects a semantic hierarchical structure as training progresses. Data samples of the same hierarchical grouping tend to exhibit greater dimensional collapse locally compared to the dataset as a whole due to sharing features in common with each other. Ideally, SSL algorithms would take advantage of this hierarchical emergence to have an additional regularization term to account for this local dimensional collapse effect. However, the construction of existing SSL algorithms does not account for this property. To address this, we propose an adaptive algorithm that performs a weighted decomposition of the denominator of the InfoNCE loss into two terms: local hierarchical and global collapse regularization respectively. This decomposition is based on an adaptive threshold that gradually lowers to reflect the emerging hierarchical structure of the representation space throughout training. It is based on an analysis of the cosine similarity distribution of samples in a batch. We demonstrate that this hierarchical emergence exploitation (HEX) approach can be integrated across a wide variety of SSL algorithms. Empirically, we show performance improvements of up to 5.6% relative improvement over baseline SSL approaches on classification accuracy on Imagenet with 100 epochs of training.
CRACKS: Crowdsourcing Resources for Analysis and Categorization of Key Subsurface faults
Aug 20, 2024



Abstract:Crowdsourcing annotations has created a paradigm shift in the availability of labeled data for machine learning. Availability of large datasets has accelerated progress in common knowledge applications involving visual and language data. However, specialized applications that require expert labels lag in data availability. One such application is fault segmentation in subsurface imaging. Detecting, tracking, and analyzing faults has broad societal implications in predicting fluid flows, earthquakes, and storing excess atmospheric CO$_2$. However, delineating faults with current practices is a labor-intensive activity that requires precise analysis of subsurface imaging data by geophysicists. In this paper, we propose the $\texttt{CRACKS}$ dataset to detect and segment faults in subsurface images by utilizing crowdsourced resources. We leverage Amazon Mechanical Turk to obtain fault delineations from sections of the Netherlands North Sea subsurface images from (i) $26$ novices who have no exposure to subsurface data and were shown a video describing and labeling faults, (ii) $8$ practitioners who have previously interacted and worked on subsurface data, (iii) one geophysicist to label $7636$ faults in the region. Note that all novices, practitioners, and the expert segment faults on the same subsurface volume with disagreements between and among the novices and practitioners. Additionally, each fault annotation is equipped with the confidence level of the annotator. The paper provides benchmarks on detecting and segmenting the expert labels, given the novice and practitioner labels. Additional details along with the dataset links and codes are available at $\href{https://alregib.ece.gatech.edu/cracks-crowdsourcing-resources-for-analysis-and-categorization-of-key-subsurface-faults/}{link}$.
Ophthalmic Biomarker Detection: Highlights from the IEEE Video and Image Processing Cup 2023 Student Competition
Aug 20, 2024



Abstract:The VIP Cup offers a unique experience to undergraduates, allowing students to work together to solve challenging, real-world problems with video and image processing techniques. In this iteration of the VIP Cup, we challenged students to balance personalization and generalization when performing biomarker detection in 3D optical coherence tomography (OCT) images. Balancing personalization and generalization is an important challenge to tackle, as the variation within OCT scans of patients between visits can be minimal while the difference in manifestation of the same disease across different patients may be substantial. The domain difference between OCT scans can arise due to pathology manifestation across patients, clinical labels, and the visit along the treatment process when the scan is taken. Hence, we provided a multimodal OCT dataset to allow teams to effectively target this challenge. Overall, this competition gave undergraduates an opportunity to learn about how artificial intelligence can be a powerful tool for the medical field, as well as the unique challenges one faces when applying machine learning to biomedical data.
Explaining Representation Learning with Perceptual Components
Jun 11, 2024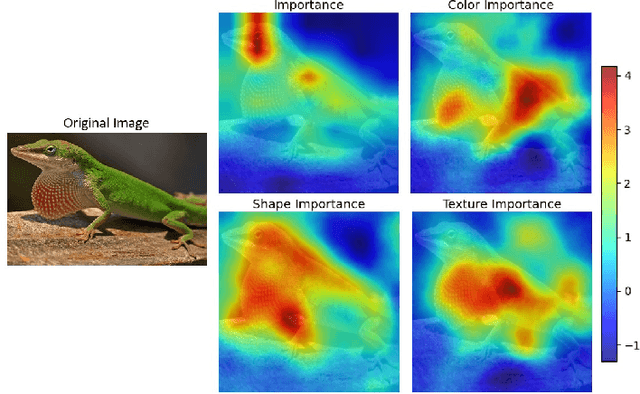

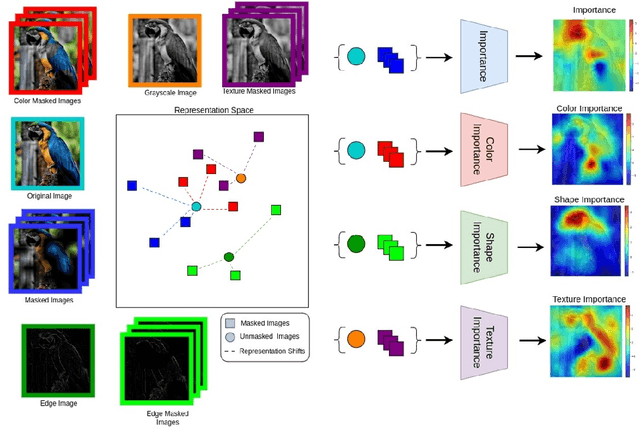
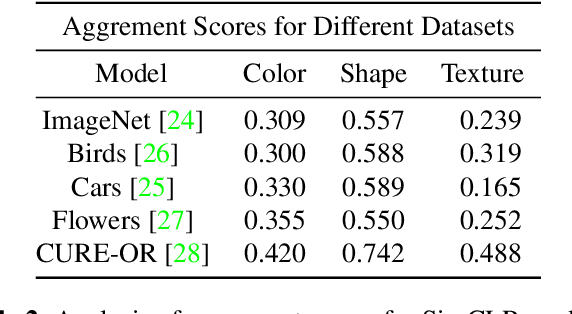
Abstract:Self-supervised models create representation spaces that lack clear semantic meaning. This interpretability problem of representations makes traditional explainability methods ineffective in this context. In this paper, we introduce a novel method to analyze representation spaces using three key perceptual components: color, shape, and texture. We employ selective masking of these components to observe changes in representations, resulting in distinct importance maps for each. In scenarios, where labels are absent, these importance maps provide more intuitive explanations as they are integral to the human visual system. Our approach enhances the interpretability of the representation space, offering explanations that resonate with human visual perception. We analyze how different training objectives create distinct representation spaces using perceptual components. Additionally, we examine the representation of images across diverse image domains, providing insights into the role of these components in different contexts.
Taxes Are All You Need: Integration of Taxonomical Hierarchy Relationships into the Contrastive Loss
Jun 10, 2024



Abstract:In this work, we propose a novel supervised contrastive loss that enables the integration of taxonomic hierarchy information during the representation learning process. A supervised contrastive loss operates by enforcing that images with the same class label (positive samples) project closer to each other than images with differing class labels (negative samples). The advantage of this approach is that it directly penalizes the structure of the representation space itself. This enables greater flexibility with respect to encoding semantic concepts. However, the standard supervised contrastive loss only enforces semantic structure based on the downstream task (i.e. the class label). In reality, the class label is only one level of a \emph{hierarchy of different semantic relationships known as a taxonomy}. For example, the class label is oftentimes the species of an animal, but between different classes there are higher order relationships such as all animals with wings being ``birds". We show that by explicitly accounting for these relationships with a weighting penalty in the contrastive loss we can out-perform the supervised contrastive loss. Additionally, we demonstrate the adaptability of the notion of a taxonomy by integrating our loss into medical and noise-based settings that show performance improvements by as much as 7%.
FOCAL: A Cost-Aware Video Dataset for Active Learning
Nov 17, 2023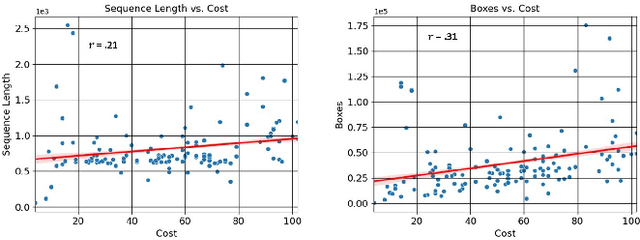
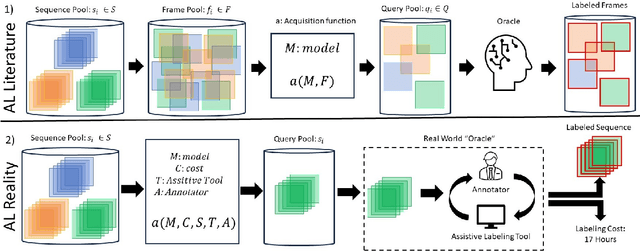
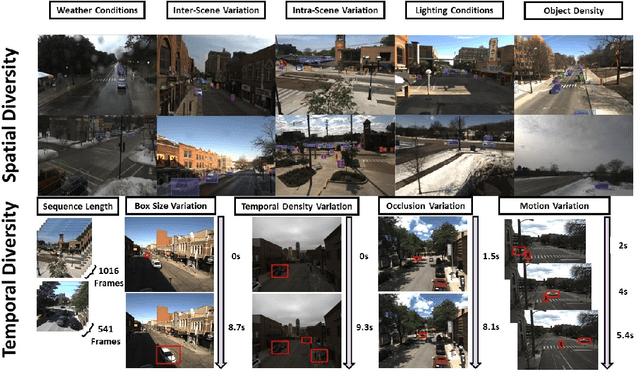
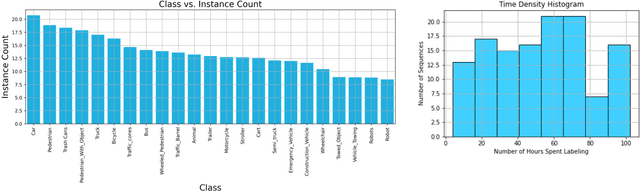
Abstract:In this paper, we introduce the FOCAL (Ford-OLIVES Collaboration on Active Learning) dataset which enables the study of the impact of annotation-cost within a video active learning setting. Annotation-cost refers to the time it takes an annotator to label and quality-assure a given video sequence. A practical motivation for active learning research is to minimize annotation-cost by selectively labeling informative samples that will maximize performance within a given budget constraint. However, previous work in video active learning lacks real-time annotation labels for accurately assessing cost minimization and instead operates under the assumption that annotation-cost scales linearly with the amount of data to annotate. This assumption does not take into account a variety of real-world confounding factors that contribute to a nonlinear cost such as the effect of an assistive labeling tool and the variety of interactions within a scene such as occluded objects, weather, and motion of objects. FOCAL addresses this discrepancy by providing real annotation-cost labels for 126 video sequences across 69 unique city scenes with a variety of weather, lighting, and seasonal conditions. We also introduce a set of conformal active learning algorithms that take advantage of the sequential structure of video data in order to achieve a better trade-off between annotation-cost and performance while also reducing floating point operations (FLOPS) overhead by at least 77.67%. We show how these approaches better reflect how annotations on videos are done in practice through a sequence selection framework. We further demonstrate the advantage of these approaches by introducing two performance-cost metrics and show that the best conformal active learning method is cheaper than the best traditional active learning method by 113 hours.
Clinical Trial Active Learning
Jul 20, 2023Abstract:This paper presents a novel approach to active learning that takes into account the non-independent and identically distributed (non-i.i.d.) structure of a clinical trial setting. There exists two types of clinical trials: retrospective and prospective. Retrospective clinical trials analyze data after treatment has been performed; prospective clinical trials collect data as treatment is ongoing. Typically, active learning approaches assume the dataset is i.i.d. when selecting training samples; however, in the case of clinical trials, treatment results in a dependency between the data collected at the current and past visits. Thus, we propose prospective active learning to overcome the limitations present in traditional active learning methods and apply it to disease detection in optical coherence tomography (OCT) images, where we condition on the time an image was collected to enforce the i.i.d. assumption. We compare our proposed method to the traditional active learning paradigm, which we refer to as retrospective in nature. We demonstrate that prospective active learning outperforms retrospective active learning in two different types of test settings.
Clinically Labeled Contrastive Learning for OCT Biomarker Classification
May 24, 2023



Abstract:This paper presents a novel positive and negative set selection strategy for contrastive learning of medical images based on labels that can be extracted from clinical data. In the medical field, there exists a variety of labels for data that serve different purposes at different stages of a diagnostic and treatment process. Clinical labels and biomarker labels are two examples. In general, clinical labels are easier to obtain in larger quantities because they are regularly collected during routine clinical care, while biomarker labels require expert analysis and interpretation to obtain. Within the field of ophthalmology, previous work has shown that clinical values exhibit correlations with biomarker structures that manifest within optical coherence tomography (OCT) scans. We exploit this relationship by using the clinical data as pseudo-labels for our data without biomarker labels in order to choose positive and negative instances for training a backbone network with a supervised contrastive loss. In this way, a backbone network learns a representation space that aligns with the clinical data distribution available. Afterwards, we fine-tune the network trained in this manner with the smaller amount of biomarker labeled data with a cross-entropy loss in order to classify these key indicators of disease directly from OCT scans. We also expand on this concept by proposing a method that uses a linear combination of clinical contrastive losses. We benchmark our methods against state of the art self-supervised methods in a novel setting with biomarkers of varying granularity. We show performance improvements by as much as 5\% in total biomarker detection AUROC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge