Kelin Xia
Topology-Aware Multiscale Mixture of Experts for Efficient Molecular Property Prediction
Jan 19, 2026Abstract:Many molecular properties depend on 3D geometry, where non-covalent interactions, stereochemical effects, and medium- to long-range forces are determined by spatial distances and angles that cannot be uniquely captured by a 2D bond graph. Yet most 3D molecular graph neural networks still rely on globally fixed neighborhood heuristics, typically defined by distance cutoffs and maximum neighbor limits, to define local message-passing neighborhoods, leading to rigid, data-agnostic interaction budgets. We propose Multiscale Interaction Mixture of Experts (MI-MoE) to adapt interaction modeling across geometric regimes. Our contributions are threefold: (1) we introduce a distance-cutoff expert ensemble that explicitly captures short-, mid-, and long-range interactions without committing to a single cutoff; (2) we design a topological gating encoder that routes inputs to experts using filtration-based descriptors, including persistent homology features, summarizing how connectivity evolves across radii; and (3) we show that MI-MoE is a plug-in module that consistently improves multiple strong 3D molecular backbones across diverse molecular and polymer property prediction benchmark datasets, covering both regression and classification tasks. These results highlight topology-aware multiscale routing as an effective principle for 3D molecular graph learning.
A roadmap for curvature-based geometric data analysis and learning
Oct 26, 2025Abstract:Geometric data analysis and learning has emerged as a distinct and rapidly developing research area, increasingly recognized for its effectiveness across diverse applications. At the heart of this field lies curvature, a powerful and interpretable concept that captures intrinsic geometric structure and underpins numerous tasks, from community detection to geometric deep learning. A wide range of discrete curvature models have been proposed for various data representations, including graphs, simplicial complexes, cubical complexes, and point clouds sampled from manifolds. These models not only provide efficient characterizations of data geometry but also constitute essential components in geometric learning frameworks. In this paper, we present the first comprehensive review of existing discrete curvature models, covering their mathematical foundations, computational formulations, and practical applications in data analysis and learning. In particular, we discuss discrete curvature from both Riemannian and metric geometry perspectives and propose a systematic pipeline for curvature-driven data analysis. We further examine the corresponding computational algorithms across different data representations, offering detailed comparisons and insights. Finally, we review state-of-the-art applications of curvature in both supervised and unsupervised learning. This survey provides a conceptual and practical roadmap for researchers to gain a better understanding of discrete curvature as a fundamental tool for geometric understanding and learning.
Rhomboid Tiling for Geometric Graph Deep Learning
May 14, 2025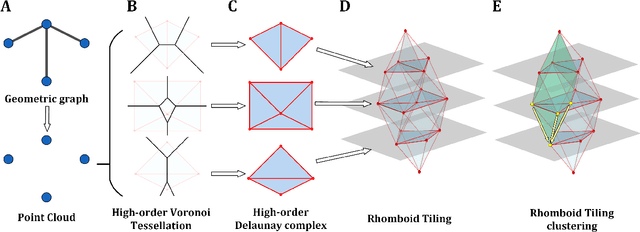
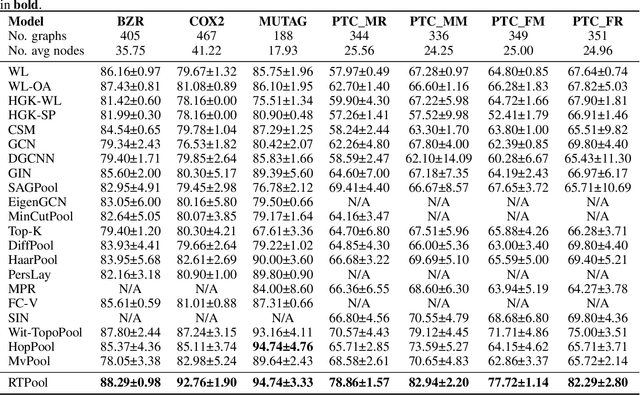
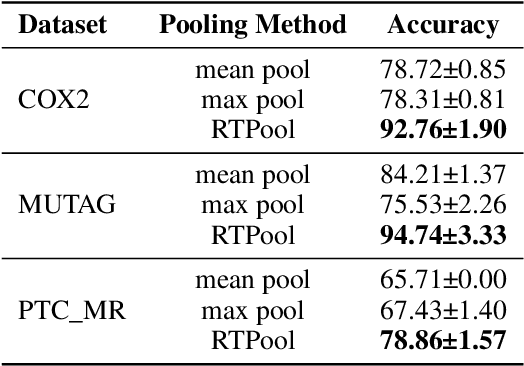
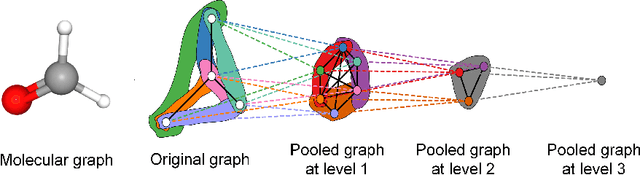
Abstract:Graph Neural Networks (GNNs) have proven effective for learning from graph-structured data through their neighborhood-based message passing framework. Many hierarchical graph clustering pooling methods modify this framework by introducing clustering-based strategies, enabling the construction of more expressive and powerful models. However, all of these message passing framework heavily rely on the connectivity structure of graphs, limiting their ability to capture the rich geometric features inherent in geometric graphs. To address this, we propose Rhomboid Tiling (RT) clustering, a novel clustering method based on the rhomboid tiling structure, which performs clustering by leveraging the complex geometric information of the data and effectively extracts its higher-order geometric structures. Moreover, we design RTPool, a hierarchical graph clustering pooling model based on RT clustering for graph classification tasks. The proposed model demonstrates superior performance, outperforming 21 state-of-the-art competitors on all the 7 benchmark datasets.
Quotient Complex Transformer (QCformer) for Perovskite Data Analysis
May 14, 2025Abstract:The discovery of novel functional materials is crucial in addressing the challenges of sustainable energy generation and climate change. Hybrid organic-inorganic perovskites (HOIPs) have gained attention for their exceptional optoelectronic properties in photovoltaics. Recently, geometric deep learning, particularly graph neural networks (GNNs), has shown strong potential in predicting material properties and guiding material design. However, traditional GNNs often struggle to capture the periodic structures and higher-order interactions prevalent in such systems. To address these limitations, we propose a novel representation based on quotient complexes (QCs) and introduce the Quotient Complex Transformer (QCformer) for material property prediction. A material structure is modeled as a quotient complex, which encodes both pairwise and many-body interactions via simplices of varying dimensions and captures material periodicity through a quotient operation. Our model leverages higher-order features defined on simplices and processes them using a simplex-based Transformer module. We pretrain QCformer on benchmark datasets such as the Materials Project and JARVIS, and fine-tune it on HOIP datasets. The results show that QCformer outperforms state-of-the-art models in HOIP property prediction, demonstrating its effectiveness. The quotient complex representation and QCformer model together contribute a powerful new tool for predictive modeling of perovskite materials.
A cohomology-based Gromov-Hausdorff metric approach for quantifying molecular similarity
Nov 21, 2024Abstract:We introduce, for the first time, a cohomology-based Gromov-Hausdorff ultrametric method to analyze 1-dimensional and higher-dimensional (co)homology groups, focusing on loops, voids, and higher-dimensional cavity structures in simplicial complexes, to address typical clustering questions arising in molecular data analysis. The Gromov-Hausdorff distance quantifies the dissimilarity between two metric spaces. In this framework, molecules are represented as simplicial complexes, and their cohomology vector spaces are computed to capture intrinsic topological invariants encoding loop and cavity structures. These vector spaces are equipped with a suitable distance measure, enabling the computation of the Gromov-Hausdorff ultrametric to evaluate structural dissimilarities. We demonstrate the methodology using organic-inorganic halide perovskite (OIHP) structures. The results highlight the effectiveness of this approach in clustering various molecular structures. By incorporating geometric information, our method provides deeper insights compared to traditional persistent homology techniques.
KA-GNN: Kolmogorov-Arnold Graph Neural Networks for Molecular Property Prediction
Oct 15, 2024Abstract:Molecular property prediction is a crucial task in the process of Artificial Intelligence-Driven Drug Discovery (AIDD). The challenge of developing models that surpass traditional non-neural network methods continues to be a vibrant area of research. This paper presents a novel graph neural network model-the Kolmogorov-Arnold Network (KAN)-based Graph Neural Network (KA-GNN), which incorporates Fourier series, specifically designed for molecular property prediction. This model maintains the high interpretability characteristic of KAN methods while being extremely efficient in computational resource usage, making it an ideal choice for deployment in resource-constrained environments. Tested and validated on seven public datasets, KA-GNN has shown significant improvements in property predictions over the existing state-of-the-art (SOTA) benchmarks.
Molecular topological deep learning for polymer property prediction
Oct 07, 2024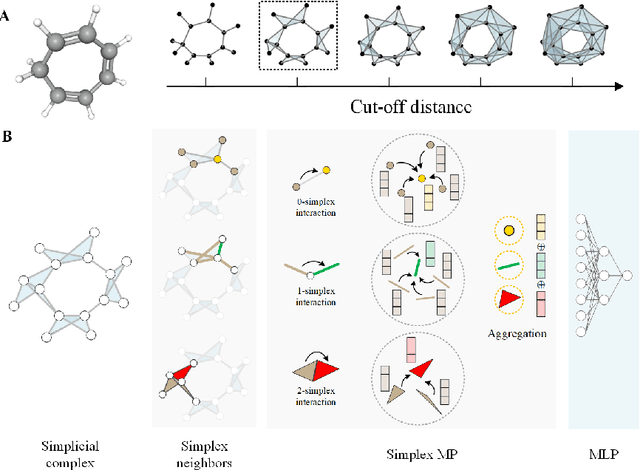
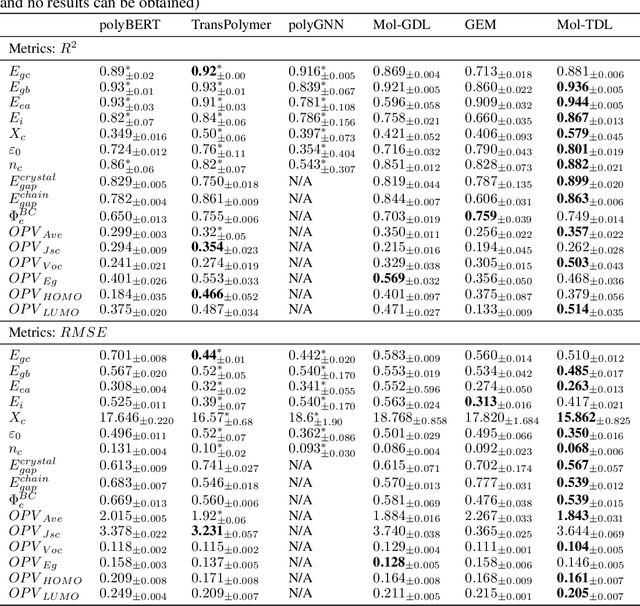
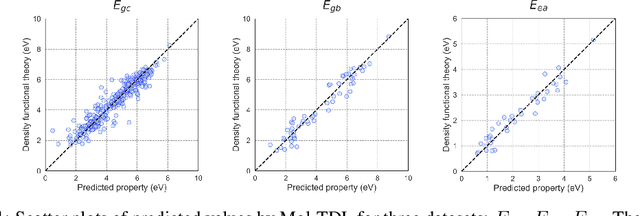
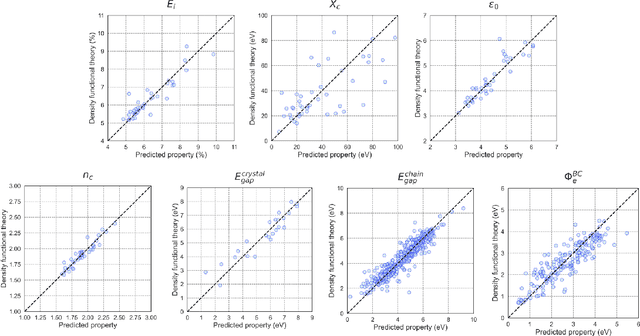
Abstract:Accurate and efficient prediction of polymer properties is of key importance for polymer design. Traditional experimental tools and density function theory (DFT)-based simulations for polymer property evaluation, are both expensive and time-consuming. Recently, a gigantic amount of graph-based molecular models have emerged and demonstrated huge potential in molecular data analysis. Even with the great progresses, these models tend to ignore the high-order and mutliscale information within the data. In this paper, we develop molecular topological deep learning (Mol-TDL) for polymer property analysis. Our Mol-TDL incorporates both high-order interactions and multiscale properties into topological deep learning architecture. The key idea is to represent polymer molecules as a series of simplicial complices at different scales and build up simplical neural networks accordingly. The aggregated information from different scales provides a more accurate prediction of polymer molecular properties.
Topology-enhanced machine learning model (Top-ML) for anticancer peptide prediction
Jul 12, 2024Abstract:Recently, therapeutic peptides have demonstrated great promise for cancer treatment. To explore powerful anticancer peptides, artificial intelligence (AI)-based approaches have been developed to systematically screen potential candidates. However, the lack of efficient featurization of peptides has become a bottleneck for these machine-learning models. In this paper, we propose a topology-enhanced machine learning model (Top-ML) for anticancer peptide prediction. Our Top-ML employs peptide topological features derived from its sequence "connection" information characterized by vector and spectral descriptors. Our Top-ML model has been validated on two widely used AntiCP 2.0 benchmark datasets and has achieved state-of-the-art performance. Our results highlight the potential of leveraging novel topology-based featurization to accelerate the identification of anticancer peptides.
Graph Neural Networks with a Distribution of Parametrized Graphs
Oct 28, 2023Abstract:Traditionally, graph neural networks have been trained using a single observed graph. However, the observed graph represents only one possible realization. In many applications, the graph may encounter uncertainties, such as having erroneous or missing edges, as well as edge weights that provide little informative value. To address these challenges and capture additional information previously absent in the observed graph, we introduce latent variables to parameterize and generate multiple graphs. We obtain the maximum likelihood estimate of the network parameters in an Expectation-Maximization (EM) framework based on the multiple graphs. Specifically, we iteratively determine the distribution of the graphs using a Markov Chain Monte Carlo (MCMC) method, incorporating the principles of PAC-Bayesian theory. Numerical experiments demonstrate improvements in performance against baseline models on node classification for heterogeneous graphs and graph regression on chemistry datasets.
Torsion Graph Neural Networks
Jun 23, 2023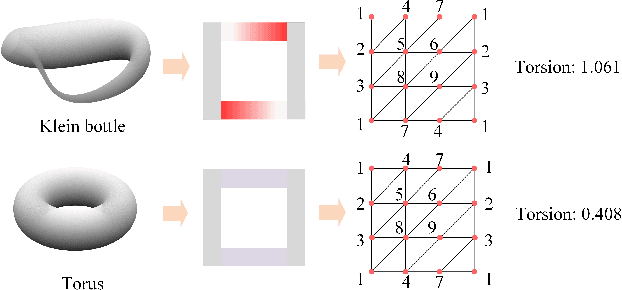
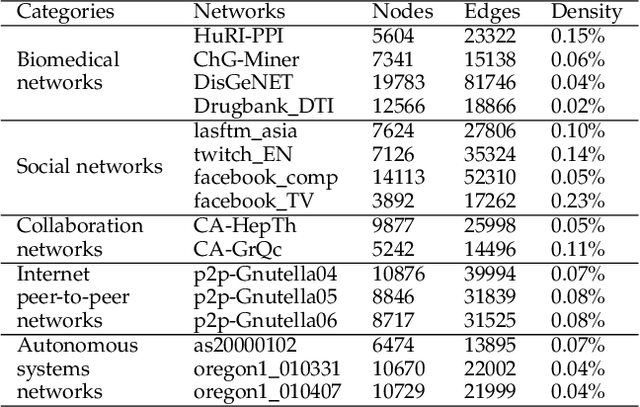
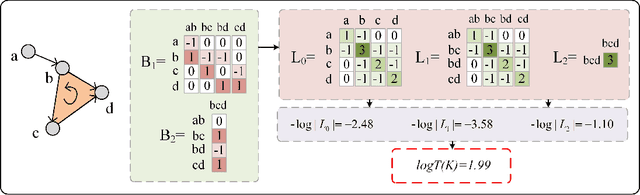
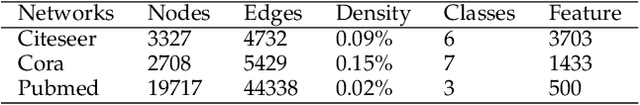
Abstract:Geometric deep learning (GDL) models have demonstrated a great potential for the analysis of non-Euclidian data. They are developed to incorporate the geometric and topological information of non-Euclidian data into the end-to-end deep learning architectures. Motivated by the recent success of discrete Ricci curvature in graph neural network (GNNs), we propose TorGNN, an analytic Torsion enhanced Graph Neural Network model. The essential idea is to characterize graph local structures with an analytic torsion based weight formula. Mathematically, analytic torsion is a topological invariant that can distinguish spaces which are homotopy equivalent but not homeomorphic. In our TorGNN, for each edge, a corresponding local simplicial complex is identified, then the analytic torsion (for this local simplicial complex) is calculated, and further used as a weight (for this edge) in message-passing process. Our TorGNN model is validated on link prediction tasks from sixteen different types of networks and node classification tasks from three types of networks. It has been found that our TorGNN can achieve superior performance on both tasks, and outperform various state-of-the-art models. This demonstrates that analytic torsion is a highly efficient topological invariant in the characterization of graph structures and can significantly boost the performance of GNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge