Ke Yuan
Divide and Conquer Self-Supervised Learning for High-Content Imaging
Mar 10, 2025Abstract:Self-supervised representation learning methods often fail to learn subtle or complex features, which can be dominated by simpler patterns which are much easier to learn. This limitation is particularly problematic in applications to science and engineering, as complex features can be critical for discovery and analysis. To address this, we introduce Split Component Embedding Registration (SpliCER), a novel architecture which splits the image into sections and distils information from each section to guide the model to learn more subtle and complex features without compromising on simpler features. SpliCER is compatible with any self-supervised loss function and can be integrated into existing methods without modification. The primary contributions of this work are as follows: i) we demonstrate that existing self-supervised methods can learn shortcut solutions when simple and complex features are both present; ii) we introduce a novel self-supervised training method, SpliCER, to overcome the limitations of existing methods, and achieve significant downstream performance improvements; iii) we demonstrate the effectiveness of SpliCER in cutting-edge medical and geospatial imaging settings. SpliCER offers a powerful new tool for representation learning, enabling models to uncover complex features which could be overlooked by other methods.
Training-Free Pretrained Model Merging
Mar 15, 2024
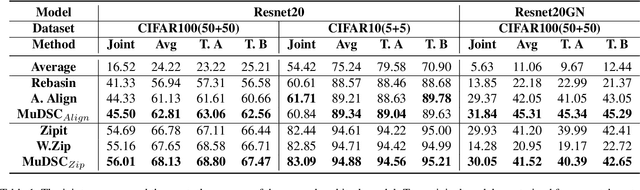
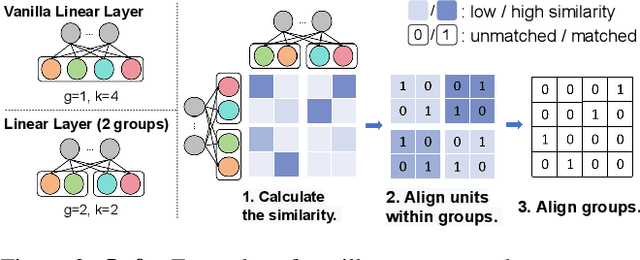
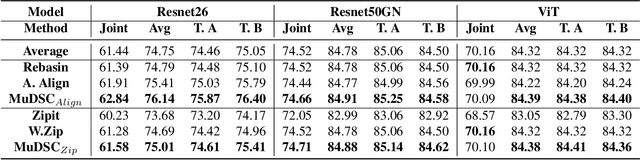
Abstract:Recently, model merging techniques have surfaced as a solution to combine multiple single-talent models into a single multi-talent model. However, previous endeavors in this field have either necessitated additional training or fine-tuning processes, or require that the models possess the same pre-trained initialization. In this work, we identify a common drawback in prior works w.r.t. the inconsistency of unit similarity in the weight space and the activation space. To address this inconsistency, we propose an innovative model merging framework, coined as merging under dual-space constraints (MuDSC). Specifically, instead of solely maximizing the objective of a single space, we advocate for the exploration of permutation matrices situated in a region with a unified high similarity in the dual space, achieved through the linear combination of activation and weight similarity matrices. In order to enhance usability, we have also incorporated adaptations for group structure, including Multi-Head Attention and Group Normalization. Comprehensive experimental comparisons demonstrate that MuDSC can significantly boost the performance of merged models with various task combinations and architectures. Furthermore, the visualization of the merged model within the multi-task loss landscape reveals that MuDSC enables the merged model to reside in the overlapping segment, featuring a unified lower loss for each task. Our code is publicly available at https://github.com/zju-vipa/training_free_model_merging.
Synthetic Privileged Information Enhances Medical Image Representation Learning
Mar 08, 2024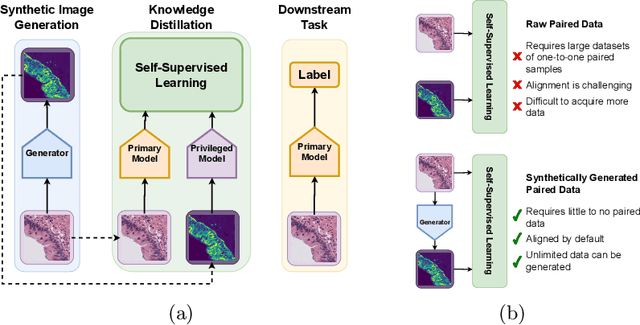
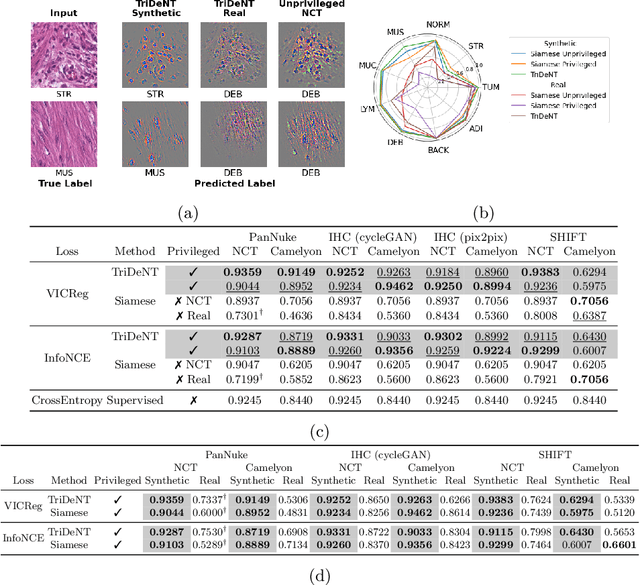
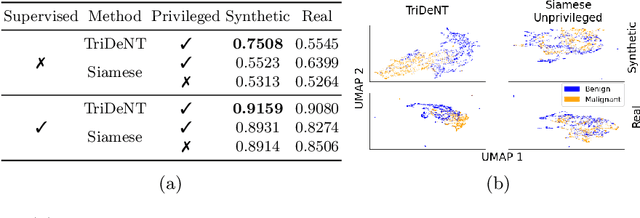
Abstract:Multimodal self-supervised representation learning has consistently proven to be a highly effective method in medical image analysis, offering strong task performance and producing biologically informed insights. However, these methods heavily rely on large, paired datasets, which is prohibitive for their use in scenarios where paired data does not exist, or there is only a small amount available. In contrast, image generation methods can work well on very small datasets, and can find mappings between unpaired datasets, meaning an effectively unlimited amount of paired synthetic data can be generated. In this work, we demonstrate that representation learning can be significantly improved by synthetically generating paired information, both compared to training on either single-modality (up to 4.4x error reduction) or authentic multi-modal paired datasets (up to 5.6x error reduction).
TriDeNT: Triple Deep Network Training for Privileged Knowledge Distillation in Histopathology
Dec 05, 2023Abstract:Computational pathology models rarely utilise data that will not be available for inference. This means most models cannot learn from highly informative data such as additional immunohistochemical (IHC) stains and spatial transcriptomics. We present TriDeNT, a novel self-supervised method for utilising privileged data that is not available during inference to improve performance. We demonstrate the efficacy of this method for a range of different paired data including immunohistochemistry, spatial transcriptomics and expert nuclei annotations. In all settings, TriDeNT outperforms other state-of-the-art methods in downstream tasks, with observed improvements of up to 101%. Furthermore, we provide qualitative and quantitative measurements of the features learned by these models and how they differ from baselines. TriDeNT offers a novel method to distil knowledge from scarce or costly data during training, to create significantly better models for routine inputs.
More From Less: Self-Supervised Knowledge Distillation for Information-Sparse Histopathology Data
Mar 19, 2023Abstract:Medical imaging technologies are generating increasingly large amounts of high-quality, information-dense data. Despite the progress, practical use of advanced imaging technologies for research and diagnosis remains limited by cost and availability, so information-sparse data such as H\&E stains are relied on in practice. The study of diseased tissue requires methods which can leverage these information-dense data to extract more value from routine, information-sparse data. Using self-supervised deep learning, we demonstrate that it is possible to distil knowledge during training from information-dense data into models which only require information-sparse data for inference. This improves downstream classification accuracy on information-sparse data, making it comparable with the fully-supervised baseline. We find substantial effects on the learned representations, and this training process identifies subtle features which otherwise go undetected. This approach enables the design of models which require only routine images, but contain insights from state-of-the-art data, allowing better use of the available resources.
Self-supervised learning unveils morphological clusters behind lung cancer types and prognosis
May 04, 2022



Abstract:Histopathological images of tumors contain abundant information about how tumors grow and how they interact with their micro-environment. Characterizing and improving our understanding of phenotypes could reveal factors related to tumor progression and their underpinning biological processes, ultimately improving diagnosis and treatment. In recent years, the field of histological deep learning applications has seen great progress, yet most of these applications focus on a supervised approach, relating tissue and associated sample annotations. Supervised approaches have their impact limited by two factors. Firstly, high-quality labels are expensive in time and effort, which makes them not easily scalable. Secondly, these methods focus on predicting annotations from histological images, fundamentally restricting the discovery of new tissue phenotypes. These limitations emphasize the importance of using new methods that can characterize tissue by the features enclosed in the image, without pre-defined annotation or supervision. We present Phenotype Representation Learning (PRL), a methodology to extract histomorphological phenotypes through self-supervised learning and community detection. PRL creates phenotype clusters by identifying tissue patterns that share common morphological and cellular features, allowing to describe whole slide images through compositional representations of cluster contributions. We used this framework to analyze histopathology slides of LUAD and LUSC lung cancer subtypes from TCGA and NYU cohorts. We show that PRL achieves a robust lung subtype prediction providing statistically relevant phenotypes for each lung subtype. We further demonstrate the significance of these phenotypes in lung adenocarcinoma overall and recurrence free survival, relating clusters with patient outcomes, cell types, grown patterns, and omic-based immune signatures.
Adversarial learning of cancer tissue representations
Aug 04, 2021



Abstract:Deep learning based analysis of histopathology images shows promise in advancing the understanding of tumor progression, tumor micro-environment, and their underpinning biological processes. So far, these approaches have focused on extracting information associated with annotations. In this work, we ask how much information can be learned from the tissue architecture itself. We present an adversarial learning model to extract feature representations of cancer tissue, without the need for manual annotations. We show that these representations are able to identify a variety of morphological characteristics across three cancer types: Breast, colon, and lung. This is supported by 1) the separation of morphologic characteristics in the latent space; 2) the ability to classify tissue type with logistic regression using latent representations, with an AUC of 0.97 and 85% accuracy, comparable to supervised deep models; 3) the ability to predict the presence of tumor in Whole Slide Images (WSIs) using multiple instance learning (MIL), achieving an AUC of 0.98 and 94% accuracy. Our results show that our model captures distinct phenotypic characteristics of real tissue samples, paving the way for further understanding of tumor progression and tumor micro-environment, and ultimately refining histopathological classification for diagnosis and treatment. The code and pretrained models are available at: https://github.com/AdalbertoCq/Adversarial-learning-of-cancer-tissue-representations
MathBERT: A Pre-Trained Model for Mathematical Formula Understanding
May 02, 2021



Abstract:Large-scale pre-trained models like BERT, have obtained a great success in various Natural Language Processing (NLP) tasks, while it is still a challenge to adapt them to the math-related tasks. Current pre-trained models neglect the structural features and the semantic correspondence between formula and its context. To address these issues, we propose a novel pre-trained model, namely \textbf{MathBERT}, which is jointly trained with mathematical formulas and their corresponding contexts. In addition, in order to further capture the semantic-level structural features of formulas, a new pre-training task is designed to predict the masked formula substructures extracted from the Operator Tree (OPT), which is the semantic structural representation of formulas. We conduct various experiments on three downstream tasks to evaluate the performance of MathBERT, including mathematical information retrieval, formula topic classification and formula headline generation. Experimental results demonstrate that MathBERT significantly outperforms existing methods on all those three tasks. Moreover, we qualitatively show that this pre-trained model effectively captures the semantic-level structural information of formulas. To the best of our knowledge, MathBERT is the first pre-trained model for mathematical formula understanding.
Automatic Description Construction for Math Expression via Topic Relation Graph
Apr 24, 2021



Abstract:Math expressions are important parts of scientific and educational documents, but some of them may be challenging for junior scholars or students to understand. Nevertheless, constructing textual descriptions for math expressions is nontrivial. In this paper, we explore the feasibility to automatically construct descriptions for math expressions. But there are two challenges that need to be addressed: 1) finding relevant documents since a math equation understanding usually requires several topics, but these topics are often explained in different documents. 2) the sparsity of the collected relevant documents making it difficult to extract reasonable descriptions. Different documents mainly focus on different topics which makes model hard to extract salient information and organize them to form a description of math expressions. To address these issues, we propose a hybrid model (MathDes) which contains two important modules: Selector and Summarizer. In the Selector, a Topic Relation Graph (TRG) is proposed to obtain the relevant documents which contain the comprehensive information of math expressions. TRG is a graph built according to the citations between expressions. In the Summarizer, a summarization model under the Integer Linear Programming (ILP) framework is proposed. This module constructs the final description with the help of a timeline that is extracted from TRG. The experimental results demonstrate that our methods are promising for this task and outperform the baselines in all aspects.
ConvMath: A Convolutional Sequence Network for Mathematical Expression Recognition
Dec 23, 2020



Abstract:Despite the recent advances in optical character recognition (OCR), mathematical expressions still face a great challenge to recognize due to their two-dimensional graphical layout. In this paper, we propose a convolutional sequence modeling network, ConvMath, which converts the mathematical expression description in an image into a LaTeX sequence in an end-to-end way. The network combines an image encoder for feature extraction and a convolutional decoder for sequence generation. Compared with other Long Short Term Memory(LSTM) based encoder-decoder models, ConvMath is entirely based on convolution, thus it is easy to perform parallel computation. Besides, the network adopts multi-layer attention mechanism in the decoder, which allows the model to align output symbols with source feature vectors automatically, and alleviates the problem of lacking coverage while training the model. The performance of ConvMath is evaluated on an open dataset named IM2LATEX-100K, including 103556 samples. The experimental results demonstrate that the proposed network achieves state-of-the-art accuracy and much better efficiency than previous methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge