John Le Quesne
Self-supervised learning unveils morphological clusters behind lung cancer types and prognosis
May 04, 2022



Abstract:Histopathological images of tumors contain abundant information about how tumors grow and how they interact with their micro-environment. Characterizing and improving our understanding of phenotypes could reveal factors related to tumor progression and their underpinning biological processes, ultimately improving diagnosis and treatment. In recent years, the field of histological deep learning applications has seen great progress, yet most of these applications focus on a supervised approach, relating tissue and associated sample annotations. Supervised approaches have their impact limited by two factors. Firstly, high-quality labels are expensive in time and effort, which makes them not easily scalable. Secondly, these methods focus on predicting annotations from histological images, fundamentally restricting the discovery of new tissue phenotypes. These limitations emphasize the importance of using new methods that can characterize tissue by the features enclosed in the image, without pre-defined annotation or supervision. We present Phenotype Representation Learning (PRL), a methodology to extract histomorphological phenotypes through self-supervised learning and community detection. PRL creates phenotype clusters by identifying tissue patterns that share common morphological and cellular features, allowing to describe whole slide images through compositional representations of cluster contributions. We used this framework to analyze histopathology slides of LUAD and LUSC lung cancer subtypes from TCGA and NYU cohorts. We show that PRL achieves a robust lung subtype prediction providing statistically relevant phenotypes for each lung subtype. We further demonstrate the significance of these phenotypes in lung adenocarcinoma overall and recurrence free survival, relating clusters with patient outcomes, cell types, grown patterns, and omic-based immune signatures.
Deconvolving convolution neural network for cell detection
Jun 18, 2018
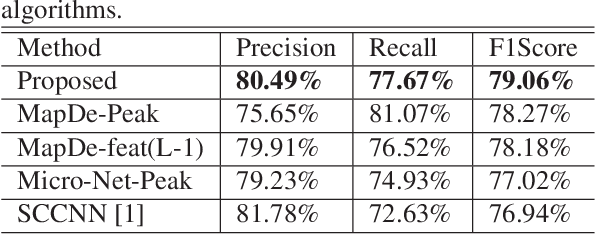
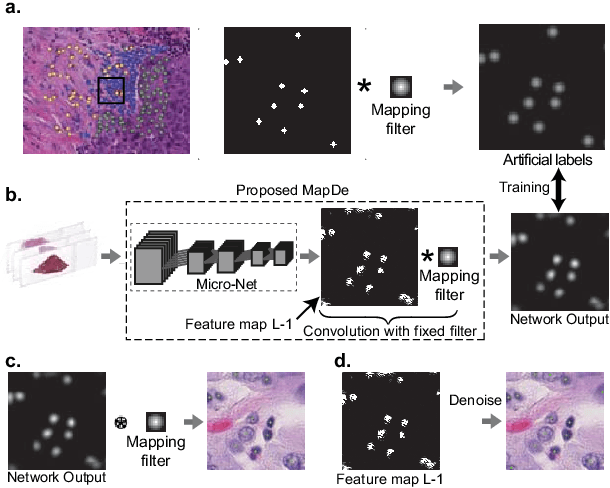
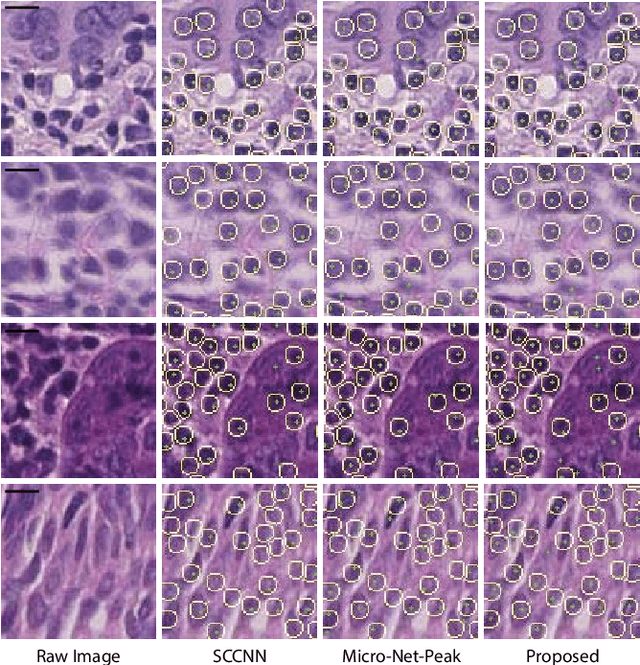
Abstract:Automatic cell detection in histology images is a challenging task due to varying size, shape and features of cells and stain variations across a large cohort. Conventional deep learning methods regress the probability of each pixel belonging to the centre of a cell followed by detection of local maxima. We present deconvolution as an alternate approach to local maxima detection. The ground truth points are convolved with a mapping filter to generate artifical labels. A convolutional neural network (CNN) is modified to convolve it's output with the same mapping filter and is trained for the mapped labels. Output of the trained CNN is then deconvolved to generate points as cell detection. We compare our method with state-of-the-art deep learning approaches where the results show that the proposed approach detects cells with comparatively high precision and F1-score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge