Adalberto Claudio Quiros
Self-supervised learning unveils morphological clusters behind lung cancer types and prognosis
May 04, 2022



Abstract:Histopathological images of tumors contain abundant information about how tumors grow and how they interact with their micro-environment. Characterizing and improving our understanding of phenotypes could reveal factors related to tumor progression and their underpinning biological processes, ultimately improving diagnosis and treatment. In recent years, the field of histological deep learning applications has seen great progress, yet most of these applications focus on a supervised approach, relating tissue and associated sample annotations. Supervised approaches have their impact limited by two factors. Firstly, high-quality labels are expensive in time and effort, which makes them not easily scalable. Secondly, these methods focus on predicting annotations from histological images, fundamentally restricting the discovery of new tissue phenotypes. These limitations emphasize the importance of using new methods that can characterize tissue by the features enclosed in the image, without pre-defined annotation or supervision. We present Phenotype Representation Learning (PRL), a methodology to extract histomorphological phenotypes through self-supervised learning and community detection. PRL creates phenotype clusters by identifying tissue patterns that share common morphological and cellular features, allowing to describe whole slide images through compositional representations of cluster contributions. We used this framework to analyze histopathology slides of LUAD and LUSC lung cancer subtypes from TCGA and NYU cohorts. We show that PRL achieves a robust lung subtype prediction providing statistically relevant phenotypes for each lung subtype. We further demonstrate the significance of these phenotypes in lung adenocarcinoma overall and recurrence free survival, relating clusters with patient outcomes, cell types, grown patterns, and omic-based immune signatures.
Adversarial learning of cancer tissue representations
Aug 04, 2021



Abstract:Deep learning based analysis of histopathology images shows promise in advancing the understanding of tumor progression, tumor micro-environment, and their underpinning biological processes. So far, these approaches have focused on extracting information associated with annotations. In this work, we ask how much information can be learned from the tissue architecture itself. We present an adversarial learning model to extract feature representations of cancer tissue, without the need for manual annotations. We show that these representations are able to identify a variety of morphological characteristics across three cancer types: Breast, colon, and lung. This is supported by 1) the separation of morphologic characteristics in the latent space; 2) the ability to classify tissue type with logistic regression using latent representations, with an AUC of 0.97 and 85% accuracy, comparable to supervised deep models; 3) the ability to predict the presence of tumor in Whole Slide Images (WSIs) using multiple instance learning (MIL), achieving an AUC of 0.98 and 94% accuracy. Our results show that our model captures distinct phenotypic characteristics of real tissue samples, paving the way for further understanding of tumor progression and tumor micro-environment, and ultimately refining histopathological classification for diagnosis and treatment. The code and pretrained models are available at: https://github.com/AdalbertoCq/Adversarial-learning-of-cancer-tissue-representations
Learning a low dimensional manifold of real cancer tissue with PathologyGAN
Apr 13, 2020



Abstract:Application of deep learning in digital pathology shows promise on improving disease diagnosis and understanding. We present a deep generative model that learns to simulate high-fidelity cancer tissue images while mapping the real images onto an interpretable low dimensional latent space. The key to the model is an encoder trained by a previously developed generative adversarial network, PathologyGAN. We study the latent space using 249K images from two breast cancer cohorts. We find that the latent space encodes morphological characteristics of tissues (e.g. patterns of cancer, lymphocytes, and stromal cells). In addition, the latent space reveals distinctly enriched clusters of tissue architectures in the high-risk patient group.
Pathology GAN: Learning deep representations of cancer tissue
Jul 04, 2019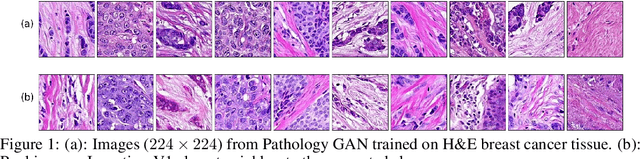

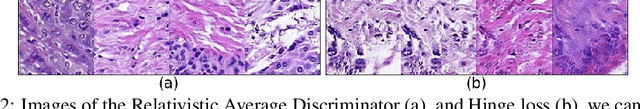
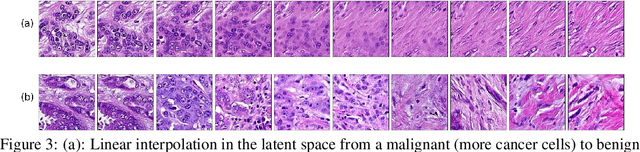
Abstract:We apply Generative Adversarial Networks (GANs) to the domain of digital pathology. Current machine learning research for digital pathology focuses on diagnosis, but we suggest a different approach and advocate that generative models could help to understand and find fundamental morphological characteristics of cancer tissue. In this paper, we develop a framework which allows GANs to capture key tissue features, and present a vision of how these could link cancer tissue and DNA in the future. To this end, we trained our model on breast cancer tissue from a medium size cohort of 526 patients, producing high fidelity images. We further study how a range of relevant GAN evaluation metrics perform on this task, and propose to evaluate synthetic images with clinically/pathologically meaningful features. Our results show that these models are able to capture key morphological characteristics that link with phenotype, such as survival time and Estrogen-receptor (ER) status. Using an Inception-V1 network as feature extraction, our models achieve a Frechet Inception Distance (FID) of 18.4. We find that using pathology meaningful features on these metrics show consistent performance, with a FID of 8.21. Furthermore, we asked two expert pathologists to distinguish our generated images from real ones, finding no significant difference between them.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge