Pathology GAN: Learning deep representations of cancer tissue
Paper and Code
Jul 04, 2019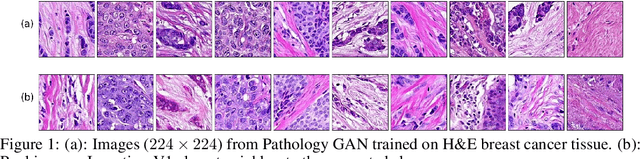

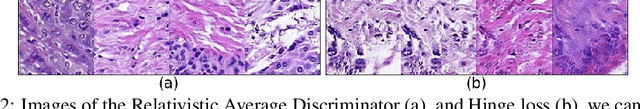
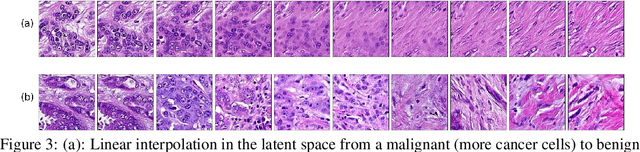
We apply Generative Adversarial Networks (GANs) to the domain of digital pathology. Current machine learning research for digital pathology focuses on diagnosis, but we suggest a different approach and advocate that generative models could help to understand and find fundamental morphological characteristics of cancer tissue. In this paper, we develop a framework which allows GANs to capture key tissue features, and present a vision of how these could link cancer tissue and DNA in the future. To this end, we trained our model on breast cancer tissue from a medium size cohort of 526 patients, producing high fidelity images. We further study how a range of relevant GAN evaluation metrics perform on this task, and propose to evaluate synthetic images with clinically/pathologically meaningful features. Our results show that these models are able to capture key morphological characteristics that link with phenotype, such as survival time and Estrogen-receptor (ER) status. Using an Inception-V1 network as feature extraction, our models achieve a Frechet Inception Distance (FID) of 18.4. We find that using pathology meaningful features on these metrics show consistent performance, with a FID of 8.21. Furthermore, we asked two expert pathologists to distinguish our generated images from real ones, finding no significant difference between them.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge