Robert Insall
Single-sample image-fusion upsampling of fluorescence lifetime images
Apr 19, 2024Abstract:Fluorescence lifetime imaging microscopy (FLIM) provides detailed information about molecular interactions and biological processes. A major bottleneck for FLIM is image resolution at high acquisition speeds, due to the engineering and signal-processing limitations of time-resolved imaging technology. Here we present single-sample image-fusion upsampling (SiSIFUS), a data-fusion approach to computational FLIM super-resolution that combines measurements from a low-resolution time-resolved detector (that measures photon arrival time) and a high-resolution camera (that measures intensity only). To solve this otherwise ill-posed inverse retrieval problem, we introduce statistically informed priors that encode local and global dependencies between the two single-sample measurements. This bypasses the risk of out-of-distribution hallucination as in traditional data-driven approaches and delivers enhanced images compared for example to standard bilinear interpolation. The general approach laid out by SiSIFUS can be applied to other image super-resolution problems where two different datasets are available.
Synthetic Privileged Information Enhances Medical Image Representation Learning
Mar 08, 2024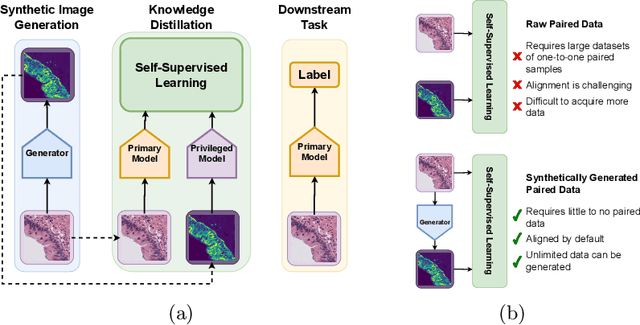
Abstract:Multimodal self-supervised representation learning has consistently proven to be a highly effective method in medical image analysis, offering strong task performance and producing biologically informed insights. However, these methods heavily rely on large, paired datasets, which is prohibitive for their use in scenarios where paired data does not exist, or there is only a small amount available. In contrast, image generation methods can work well on very small datasets, and can find mappings between unpaired datasets, meaning an effectively unlimited amount of paired synthetic data can be generated. In this work, we demonstrate that representation learning can be significantly improved by synthetically generating paired information, both compared to training on either single-modality (up to 4.4x error reduction) or authentic multi-modal paired datasets (up to 5.6x error reduction).
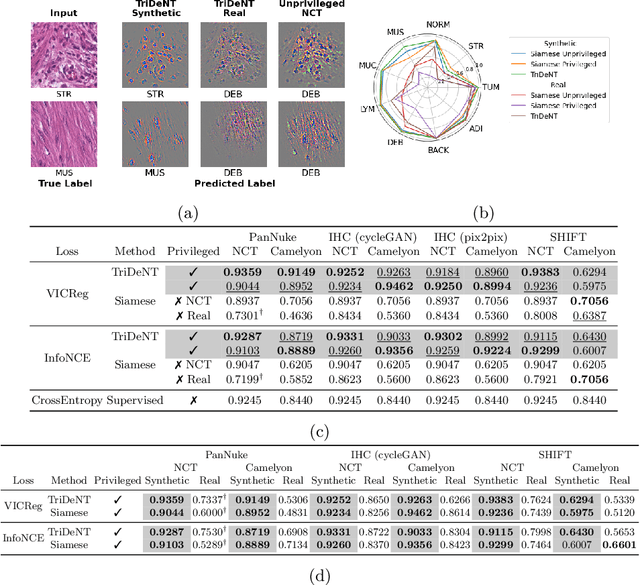
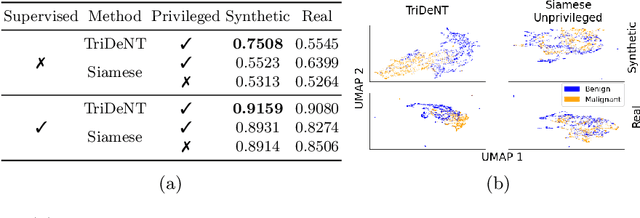
TriDeNT: Triple Deep Network Training for Privileged Knowledge Distillation in Histopathology
Dec 05, 2023Abstract:Computational pathology models rarely utilise data that will not be available for inference. This means most models cannot learn from highly informative data such as additional immunohistochemical (IHC) stains and spatial transcriptomics. We present TriDeNT, a novel self-supervised method for utilising privileged data that is not available during inference to improve performance. We demonstrate the efficacy of this method for a range of different paired data including immunohistochemistry, spatial transcriptomics and expert nuclei annotations. In all settings, TriDeNT outperforms other state-of-the-art methods in downstream tasks, with observed improvements of up to 101%. Furthermore, we provide qualitative and quantitative measurements of the features learned by these models and how they differ from baselines. TriDeNT offers a novel method to distil knowledge from scarce or costly data during training, to create significantly better models for routine inputs.
More From Less: Self-Supervised Knowledge Distillation for Information-Sparse Histopathology Data
Mar 19, 2023Abstract:Medical imaging technologies are generating increasingly large amounts of high-quality, information-dense data. Despite the progress, practical use of advanced imaging technologies for research and diagnosis remains limited by cost and availability, so information-sparse data such as H\&E stains are relied on in practice. The study of diseased tissue requires methods which can leverage these information-dense data to extract more value from routine, information-sparse data. Using self-supervised deep learning, we demonstrate that it is possible to distil knowledge during training from information-dense data into models which only require information-sparse data for inference. This improves downstream classification accuracy on information-sparse data, making it comparable with the fully-supervised baseline. We find substantial effects on the learned representations, and this training process identifies subtle features which otherwise go undetected. This approach enables the design of models which require only routine images, but contain insights from state-of-the-art data, allowing better use of the available resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge