Karl Øyvind Mikalsen
The Norwegian Centre for Clinical Artificial Intelligence
Natural Language Processing for Electronic Health Records in Scandinavian Languages: Norwegian, Swedish, and Danish
Mar 24, 2025



Abstract:Background: Clinical natural language processing (NLP) refers to the use of computational methods for extracting, processing, and analyzing unstructured clinical text data, and holds a huge potential to transform healthcare in various clinical tasks. Objective: The study aims to perform a systematic review to comprehensively assess and analyze the state-of-the-art NLP methods for the mainland Scandinavian clinical text. Method: A literature search was conducted in various online databases including PubMed, ScienceDirect, Google Scholar, ACM digital library, and IEEE Xplore between December 2022 and February 2024. Further, relevant references to the included articles were also used to solidify our search. The final pool includes articles that conducted clinical NLP in the mainland Scandinavian languages and were published in English between 2010 and 2024. Results: Out of the 113 articles, 18% (n=21) focus on Norwegian clinical text, 64% (n=72) on Swedish, 10% (n=11) on Danish, and 8% (n=9) focus on more than one language. Generally, the review identified positive developments across the region despite some observable gaps and disparities between the languages. There are substantial disparities in the level of adoption of transformer-based models. In essential tasks such as de-identification, there is significantly less research activity focusing on Norwegian and Danish compared to Swedish text. Further, the review identified a low level of sharing resources such as data, experimentation code, pre-trained models, and rate of adaptation and transfer learning in the region. Conclusion: The review presented a comprehensive assessment of the state-of-the-art Clinical NLP for electronic health records (EHR) text in mainland Scandinavian languages and, highlighted the potential barriers and challenges that hinder the rapid advancement of the field in the region.
Artificial intelligence to improve clinical coding practice in Scandinavia: a crossover randomized controlled trial
Oct 31, 2024Abstract:\textbf{Trial design} Crossover randomized controlled trial. \textbf{Methods} An AI tool, Easy-ICD, was developed to assist clinical coders and was tested for improving both accuracy and time in a user study in Norway and Sweden. Participants were randomly assigned to two groups, and crossed over between coding complex (longer) texts versus simple (shorter) texts, while using our tool versus not using our tool. \textbf{Results} Based on Mann-Whitney U test, the median coding time difference for complex clinical text sequences was 123 seconds (\emph{P}\textless.001, 95\% CI: 81 to 164), representing a 46\% reduction in median coding time when our tool is used. There was no significant time difference for simpler text sequences. For coding accuracy, the improvement we noted for both complex and simple texts was not significant. \textbf{Conclusions} This study demonstrates the potential of AI to transform common tasks in clinical workflows, with ostensible positive impacts on work efficiencies for complex clinical coding tasks. Further studies within hospital workflows are required before these presumed impacts can be more clearly understood.
Approaching adverse event detection utilizing transformers on clinical time-series
Nov 15, 2023Abstract:Patients being admitted to a hospital will most often be associated with a certain clinical development during their stay. However, there is always a risk of patients being subject to the wrong diagnosis or to a certain treatment not pertaining to the desired effect, potentially leading to adverse events. Our research aims to develop an anomaly detection system for identifying deviations from expected clinical trajectories. To address this goal we analyzed 16 months of vital sign recordings obtained from the Nordland Hospital Trust (NHT). We employed an self-supervised framework based on the STraTS transformer architecture to represent the time series data in a latent space. These representations were then subjected to various clustering techniques to explore potential patient phenotypes based on their clinical progress. While our preliminary results from this ongoing research are promising, they underscore the importance of enhancing the dataset with additional demographic information from patients. This additional data will be crucial for a more comprehensive evaluation of the method's performance.
A clinically motivated self-supervised approach for content-based image retrieval of CT liver images
Jul 11, 2022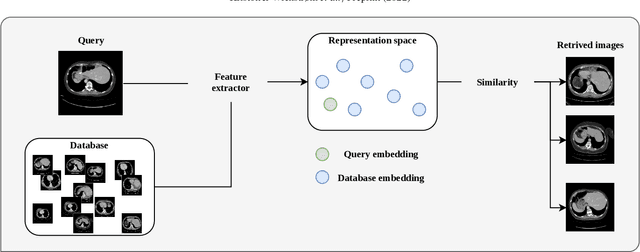
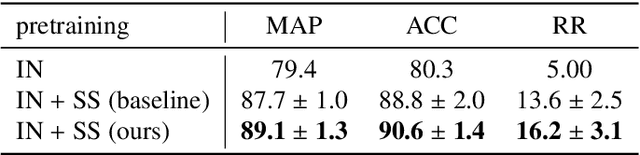
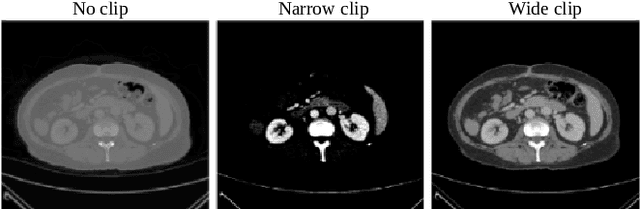
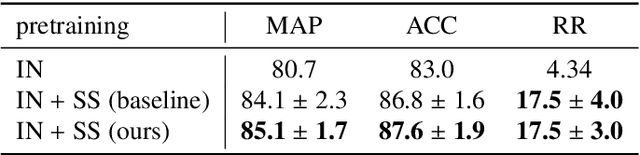
Abstract:Deep learning-based approaches for content-based image retrieval (CBIR) of CT liver images is an active field of research, but suffers from some critical limitations. First, they are heavily reliant on labeled data, which can be challenging and costly to acquire. Second, they lack transparency and explainability, which limits the trustworthiness of deep CBIR systems. We address these limitations by (1) proposing a self-supervised learning framework that incorporates domain-knowledge into the training procedure and (2) providing the first representation learning explainability analysis in the context of CBIR of CT liver images. Results demonstrate improved performance compared to the standard self-supervised approach across several metrics, as well as improved generalisation across datasets. Further, we conduct the first representation learning explainability analysis in the context of CBIR, which reveals new insights into the feature extraction process. Lastly, we perform a case study with cross-examination CBIR that demonstrates the usability of our proposed framework. We believe that our proposed framework could play a vital role in creating trustworthy deep CBIR systems that can successfully take advantage of unlabeled data.
The Kernelized Taylor Diagram
May 18, 2022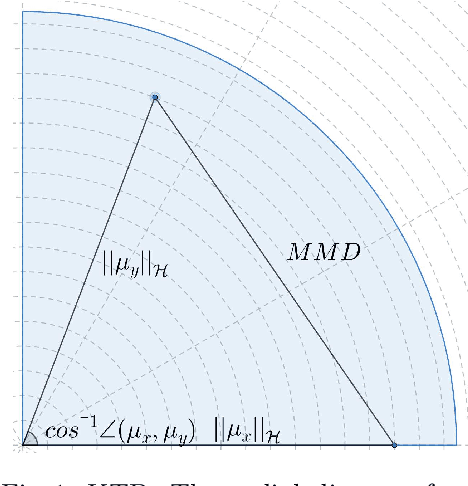
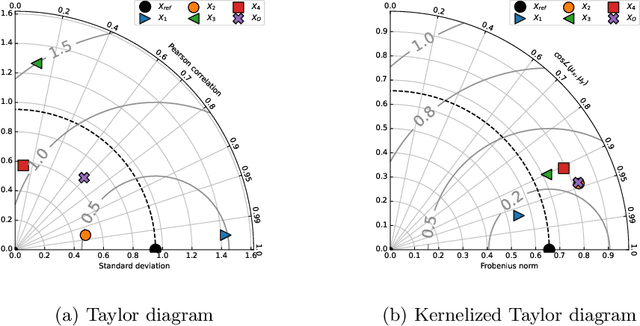
Abstract:This paper presents the kernelized Taylor diagram, a graphical framework for visualizing similarities between data populations. The kernelized Taylor diagram builds on the widely used Taylor diagram, which is used to visualize similarities between populations. However, the Taylor diagram has several limitations such as not capturing non-linear relationships and sensitivity to outliers. To address such limitations, we propose the kernelized Taylor diagram. Our proposed kernelized Taylor diagram is capable of visualizing similarities between populations with minimal assumptions of the data distributions. The kernelized Taylor diagram relates the maximum mean discrepancy and the kernel mean embedding in a single diagram, a construction that, to the best of our knowledge, have not been devised prior to this work. We believe that the kernelized Taylor diagram can be a valuable tool in data visualization.
Mixing Up Contrastive Learning: Self-Supervised Representation Learning for Time Series
Mar 17, 2022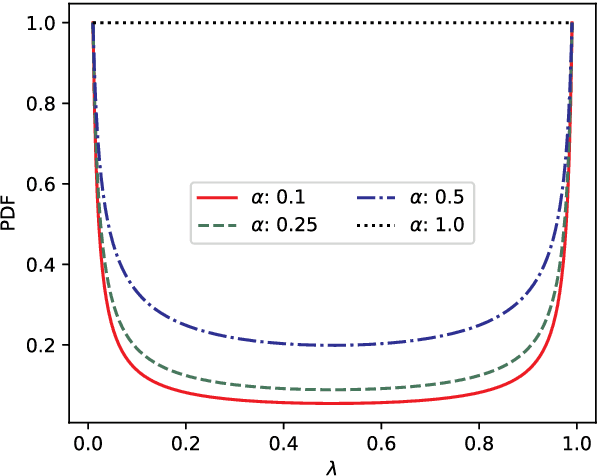
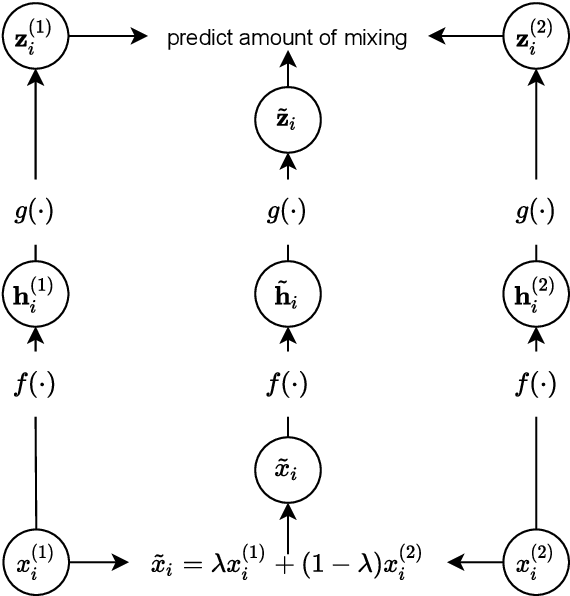
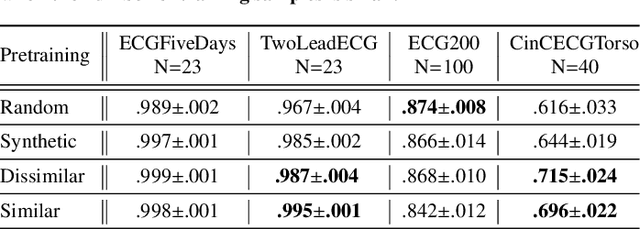
Abstract:The lack of labeled data is a key challenge for learning useful representation from time series data. However, an unsupervised representation framework that is capable of producing high quality representations could be of great value. It is key to enabling transfer learning, which is especially beneficial for medical applications, where there is an abundance of data but labeling is costly and time consuming. We propose an unsupervised contrastive learning framework that is motivated from the perspective of label smoothing. The proposed approach uses a novel contrastive loss that naturally exploits a data augmentation scheme in which new samples are generated by mixing two data samples with a mixing component. The task in the proposed framework is to predict the mixing component, which is utilized as soft targets in the loss function. Experiments demonstrate the framework's superior performance compared to other representation learning approaches on both univariate and multivariate time series and illustrate its benefits for transfer learning for clinical time series.
RELAX: Representation Learning Explainability
Dec 19, 2021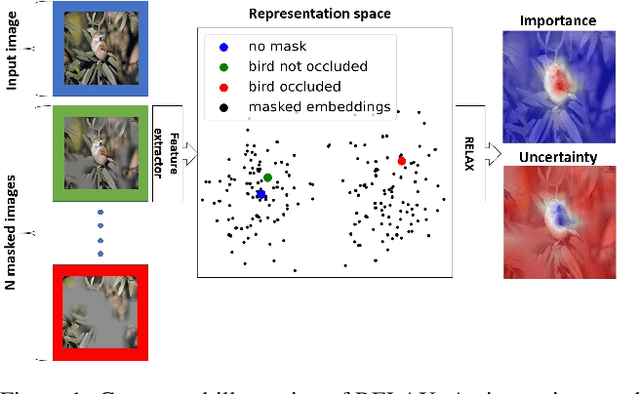

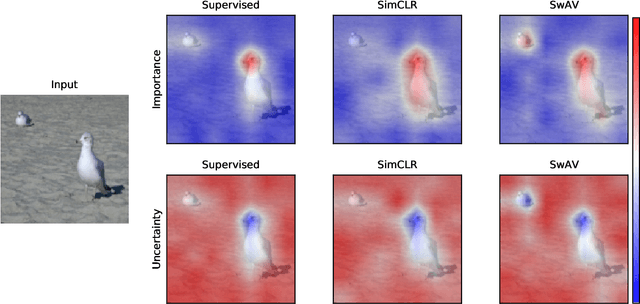
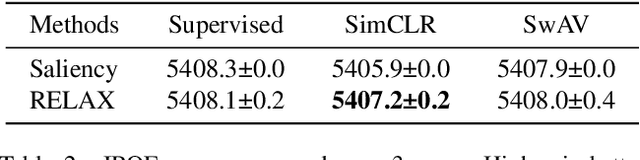
Abstract:Despite the significant improvements that representation learning via self-supervision has led to when learning from unlabeled data, no methods exist that explain what influences the learned representation. We address this need through our proposed approach, RELAX, which is the first approach for attribution-based explanations of representations. Our approach can also model the uncertainty in its explanations, which is essential to produce trustworthy explanations. RELAX explains representations by measuring similarities in the representation space between an input and masked out versions of itself, providing intuitive explanations and significantly outperforming the gradient-based baseline. We provide theoretical interpretations of RELAX and conduct a novel analysis of feature extractors trained using supervised and unsupervised learning, providing insights into different learning strategies. Finally, we illustrate the usability of RELAX in multi-view clustering and highlight that incorporating uncertainty can be essential for providing low-complexity explanations, taking a crucial step towards explaining representations.
On the Use of Time Series Kernel and Dimensionality Reduction to Identify the Acquisition of Antimicrobial Multidrug Resistance in the Intensive Care Unit
Jul 07, 2021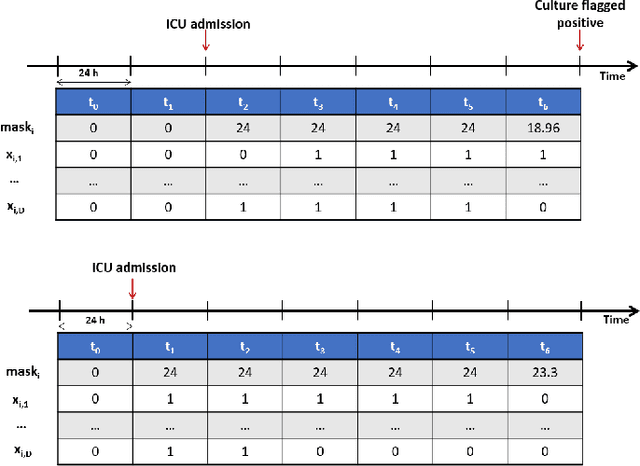
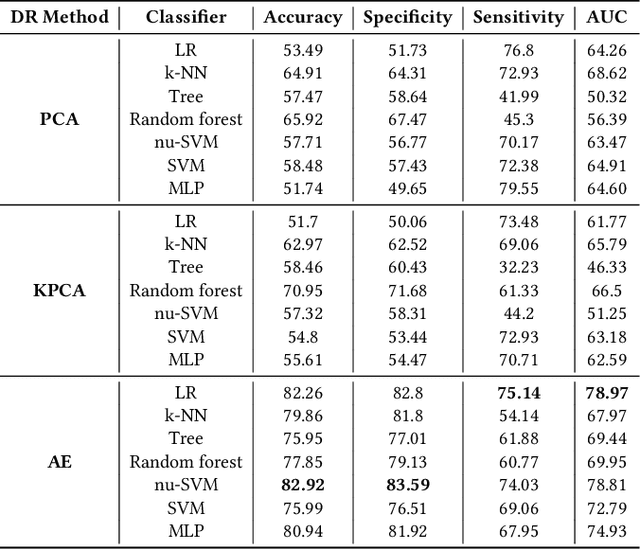
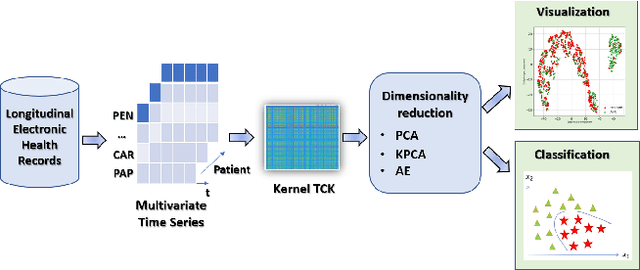
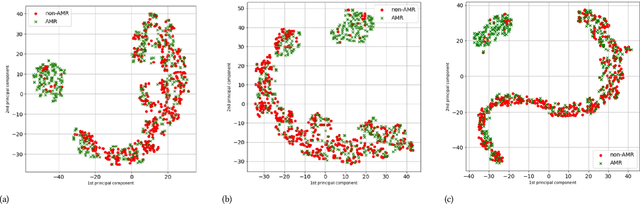
Abstract:The acquisition of Antimicrobial Multidrug Resistance (AMR) in patients admitted to the Intensive Care Units (ICU) is a major global concern. This study analyses data in the form of multivariate time series (MTS) from 3476 patients recorded at the ICU of University Hospital of Fuenlabrada (Madrid) from 2004 to 2020. 18\% of the patients acquired AMR during their stay in the ICU. The goal of this paper is an early prediction of the development of AMR. Towards that end, we leverage the time-series cluster kernel (TCK) to learn similarities between MTS. To evaluate the effectiveness of TCK as a kernel, we applied several dimensionality reduction techniques for visualization and classification tasks. The experimental results show that TCK allows identifying a group of patients that acquire the AMR during the first 48 hours of their ICU stay, and it also provides good classification capabilities.
Uncertainty-Aware Deep Ensembles for Reliable and Explainable Predictions of Clinical Time Series
Oct 16, 2020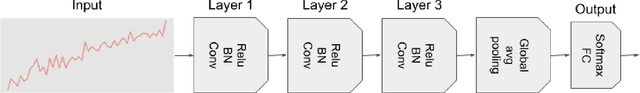
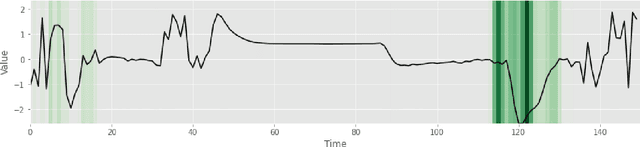
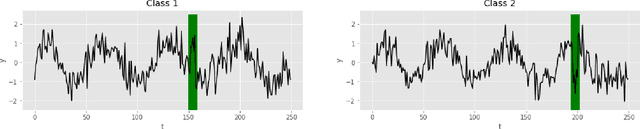
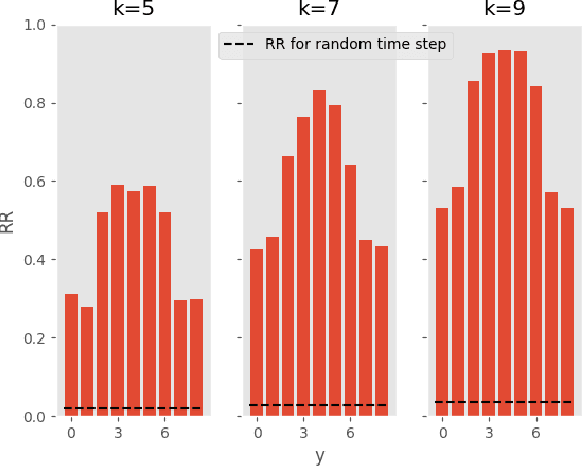
Abstract:Deep learning-based support systems have demonstrated encouraging results in numerous clinical applications involving the processing of time series data. While such systems often are very accurate, they have no inherent mechanism for explaining what influenced the predictions, which is critical for clinical tasks. However, existing explainability techniques lack an important component for trustworthy and reliable decision support, namely a notion of uncertainty. In this paper, we address this lack of uncertainty by proposing a deep ensemble approach where a collection of DNNs are trained independently. A measure of uncertainty in the relevance scores is computed by taking the standard deviation across the relevance scores produced by each model in the ensemble, which in turn is used to make the explanations more reliable. The class activation mapping method is used to assign a relevance score for each time step in the time series. Results demonstrate that the proposed ensemble is more accurate in locating relevant time steps and is more consistent across random initializations, thus making the model more trustworthy. The proposed methodology paves the way for constructing trustworthy and dependable support systems for processing clinical time series for healthcare related tasks.
A Kernel to Exploit Informative Missingness in Multivariate Time Series from EHRs
Feb 27, 2020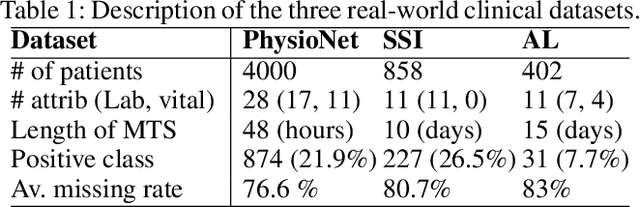
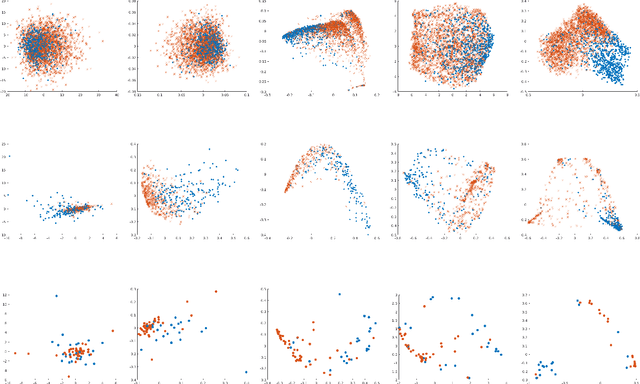
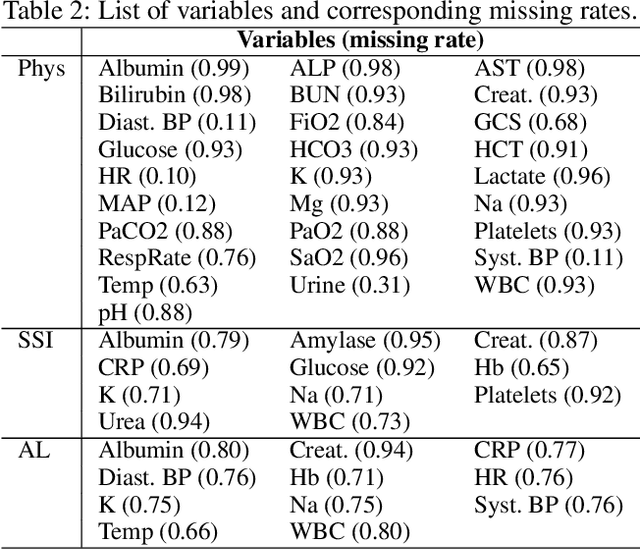
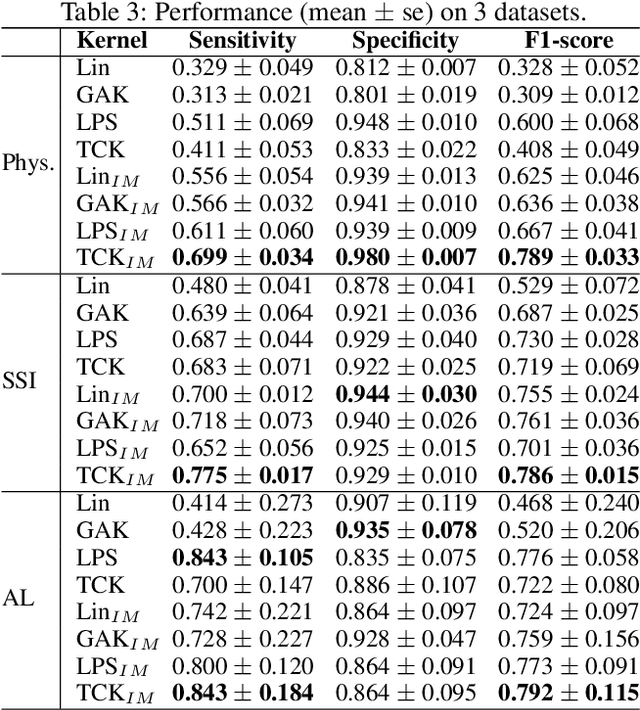
Abstract:A large fraction of the electronic health records (EHRs) consists of clinical measurements collected over time, such as lab tests and vital signs, which provide important information about a patient's health status. These sequences of clinical measurements are naturally represented as time series, characterized by multiple variables and large amounts of missing data, which complicate the analysis. In this work, we propose a novel kernel which is capable of exploiting both the information from the observed values as well the information hidden in the missing patterns in multivariate time series (MTS) originating e.g. from EHRs. The kernel, called TCK$_{IM}$, is designed using an ensemble learning strategy in which the base models are novel mixed mode Bayesian mixture models which can effectively exploit informative missingness without having to resort to imputation methods. Moreover, the ensemble approach ensures robustness to hyperparameters and therefore TCK$_{IM}$ is particularly well suited if there is a lack of labels - a known challenge in medical applications. Experiments on three real-world clinical datasets demonstrate the effectiveness of the proposed kernel.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge