Arthur Revhaug
Uncertainty-Aware Deep Ensembles for Reliable and Explainable Predictions of Clinical Time Series
Oct 16, 2020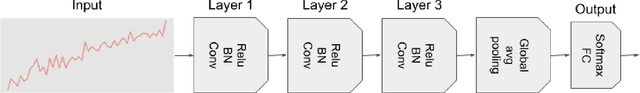
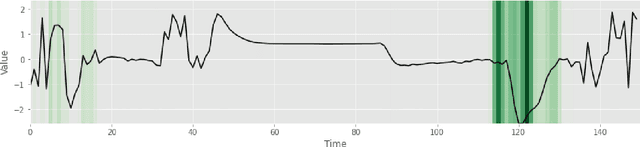
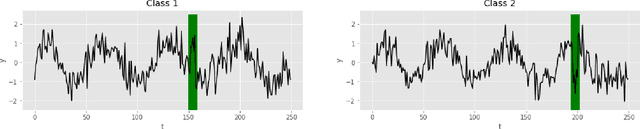
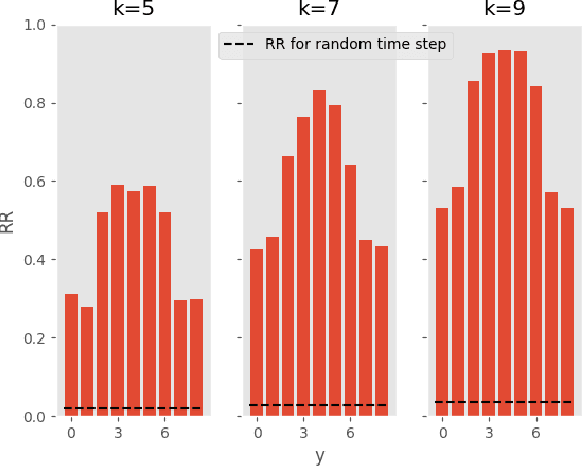
Abstract:Deep learning-based support systems have demonstrated encouraging results in numerous clinical applications involving the processing of time series data. While such systems often are very accurate, they have no inherent mechanism for explaining what influenced the predictions, which is critical for clinical tasks. However, existing explainability techniques lack an important component for trustworthy and reliable decision support, namely a notion of uncertainty. In this paper, we address this lack of uncertainty by proposing a deep ensemble approach where a collection of DNNs are trained independently. A measure of uncertainty in the relevance scores is computed by taking the standard deviation across the relevance scores produced by each model in the ensemble, which in turn is used to make the explanations more reliable. The class activation mapping method is used to assign a relevance score for each time step in the time series. Results demonstrate that the proposed ensemble is more accurate in locating relevant time steps and is more consistent across random initializations, thus making the model more trustworthy. The proposed methodology paves the way for constructing trustworthy and dependable support systems for processing clinical time series for healthcare related tasks.
Time series cluster kernels to exploit informative missingness and incomplete label information
Jul 10, 2019
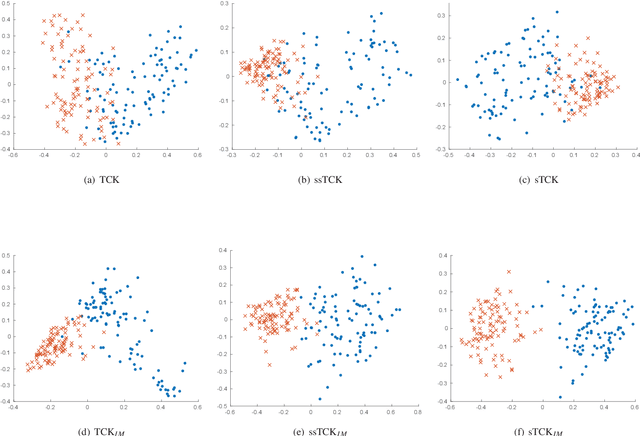
Abstract:The time series cluster kernel (TCK) provides a powerful tool for analysing multivariate time series subject to missing data. TCK is designed using an ensemble learning approach in which Bayesian mixture models form the base models. Because of the Bayesian approach, TCK can naturally deal with missing values without resorting to imputation and the ensemble strategy ensures robustness to hyperparameters, making it particularly well suited for unsupervised learning. However, TCK assumes missing at random and that the underlying missingness mechanism is ignorable, i.e. uninformative, an assumption that does not hold in many real-world applications, such as e.g. medicine. To overcome this limitation, we present a kernel capable of exploiting the potentially rich information in the missing values and patterns, as well as the information from the observed data. In our approach, we create a representation of the missing pattern, which is incorporated into mixed mode mixture models in such a way that the information provided by the missing patterns is effectively exploited. Moreover, we also propose a semi-supervised kernel, capable of taking advantage of incomplete label information to learn more accurate similarities. Experiments on benchmark data, as well as a real-world case study of patients described by longitudinal electronic health record data who potentially suffer from hospital-acquired infections, demonstrate the effectiveness of the proposed methods.
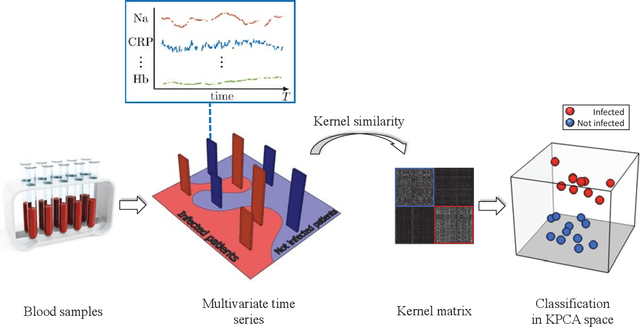
An Unsupervised Multivariate Time Series Kernel Approach for Identifying Patients with Surgical Site Infection from Blood Samples
Mar 21, 2018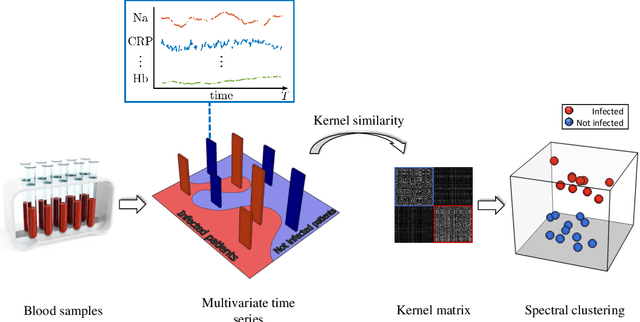
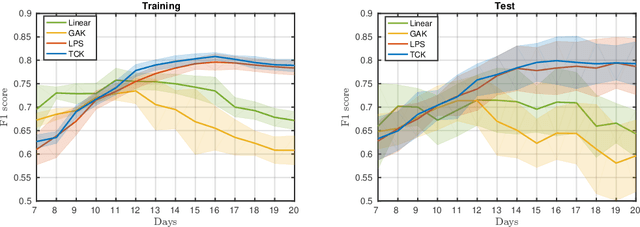
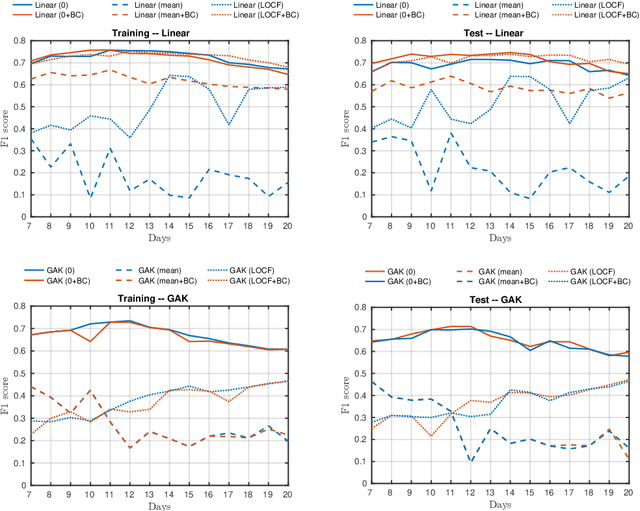
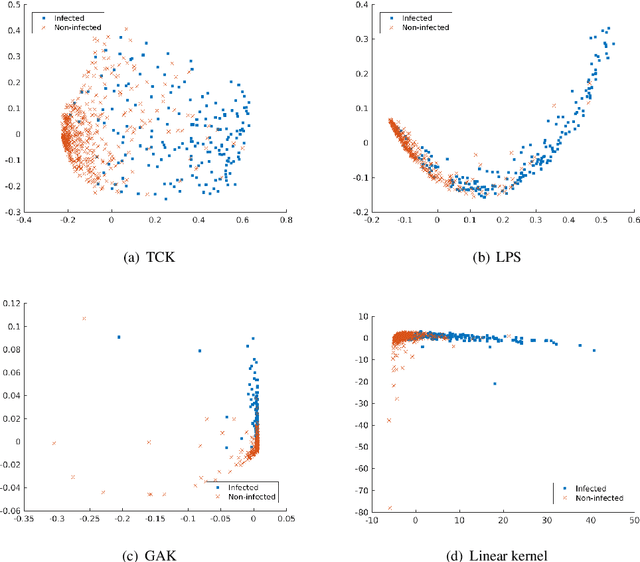
Abstract:A large fraction of the electronic health records consists of clinical measurements collected over time, such as blood tests, which provide important information about the health status of a patient. These sequences of clinical measurements are naturally represented as time series, characterized by multiple variables and the presence of missing data, which complicate analysis. In this work, we propose a surgical site infection detection framework for patients undergoing colorectal cancer surgery that is completely unsupervised, hence alleviating the problem of getting access to labelled training data. The framework is based on powerful kernels for multivariate time series that account for missing data when computing similarities. Our approach show superior performance compared to baselines that have to resort to imputation techniques and performs comparable to a supervised classification baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge