Junyu Yan
SWiFT: Soft-Mask Weight Fine-tuning for Bias Mitigation
Aug 26, 2025Abstract:Recent studies have shown that Machine Learning (ML) models can exhibit bias in real-world scenarios, posing significant challenges in ethically sensitive domains such as healthcare. Such bias can negatively affect model fairness, model generalization abilities and further risks amplifying social discrimination. There is a need to remove biases from trained models. Existing debiasing approaches often necessitate access to original training data and need extensive model retraining; they also typically exhibit trade-offs between model fairness and discriminative performance. To address these challenges, we propose Soft-Mask Weight Fine-Tuning (SWiFT), a debiasing framework that efficiently improves fairness while preserving discriminative performance with much less debiasing costs. Notably, SWiFT requires only a small external dataset and only a few epochs of model fine-tuning. The idea behind SWiFT is to first find the relative, and yet distinct, contributions of model parameters to both bias and predictive performance. Then, a two-step fine-tuning process updates each parameter with different gradient flows defined by its contribution. Extensive experiments with three bias sensitive attributes (gender, skin tone, and age) across four dermatological and two chest X-ray datasets demonstrate that SWiFT can consistently reduce model bias while achieving competitive or even superior diagnostic accuracy under common fairness and accuracy metrics, compared to the state-of-the-art. Specifically, we demonstrate improved model generalization ability as evidenced by superior performance on several out-of-distribution (OOD) datasets.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:015
Anatomy-Grounded Weakly Supervised Prompt Tuning for Chest X-ray Latent Diffusion Models
Jun 12, 2025Abstract:Latent Diffusion Models have shown remarkable results in text-guided image synthesis in recent years. In the domain of natural (RGB) images, recent works have shown that such models can be adapted to various vision-language downstream tasks with little to no supervision involved. On the contrary, text-to-image Latent Diffusion Models remain relatively underexplored in the field of medical imaging, primarily due to limited data availability (e.g., due to privacy concerns). In this work, focusing on the chest X-ray modality, we first demonstrate that a standard text-conditioned Latent Diffusion Model has not learned to align clinically relevant information in free-text radiology reports with the corresponding areas of the given scan. Then, to alleviate this issue, we propose a fine-tuning framework to improve multi-modal alignment in a pre-trained model such that it can be efficiently repurposed for downstream tasks such as phrase grounding. Our method sets a new state-of-the-art on a standard benchmark dataset (MS-CXR), while also exhibiting robust performance on out-of-distribution data (VinDr-CXR). Our code will be made publicly available.
BMFT: Achieving Fairness via Bias-based Weight Masking Fine-tuning
Aug 13, 2024Abstract:Developing models with robust group fairness properties is paramount, particularly in ethically sensitive domains such as medical diagnosis. Recent approaches to achieving fairness in machine learning require a substantial amount of training data and depend on model retraining, which may not be practical in real-world scenarios. To mitigate these challenges, we propose Bias-based Weight Masking Fine-Tuning (BMFT), a novel post-processing method that enhances the fairness of a trained model in significantly fewer epochs without requiring access to the original training data. BMFT produces a mask over model parameters, which efficiently identifies the weights contributing the most towards biased predictions. Furthermore, we propose a two-step debiasing strategy, wherein the feature extractor undergoes initial fine-tuning on the identified bias-influenced weights, succeeded by a fine-tuning phase on a reinitialised classification layer to uphold discriminative performance. Extensive experiments across four dermatological datasets and two sensitive attributes demonstrate that BMFT outperforms existing state-of-the-art (SOTA) techniques in both diagnostic accuracy and fairness metrics. Our findings underscore the efficacy and robustness of BMFT in advancing fairness across various out-of-distribution (OOD) settings. Our code is available at: https://github.com/vios-s/BMFT
New Interaction Paradigm for Complex EDA Software Leveraging GPT
Jul 27, 2023



Abstract:In the rapidly growing field of electronic design automation (EDA), professional software such as KiCad, Cadence , and Altium Designer provide increasingly extensive design functionalities. However, the intricate command structure and high learning curve create a barrier, particularly for novice printed circuit board (PCB) designers. This results in difficulties in selecting appropriate functions or plugins for varying design purposes, compounded by the lack of intuitive learning methods beyond traditional documentation, videos, and online forums. To address this challenge, an artificial intelligence (AI) interaction assist plugin for EDA software named SmartonAl is developed here, also KiCad is taken as the first example. SmartonAI is inspired by the HuggingGPT framework and employs large language models, such as GPT and BERT, to facilitate task planning and execution. On receiving a designer request, SmartonAI conducts a task breakdown and efficiently executes relevant subtasks, such as analysis of help documentation paragraphs and execution of different plugins, along with leveraging the built-in schematic and PCB manipulation functions in both SmartonAl itself and software. Our preliminary results demonstrate that SmartonAI can significantly streamline the PCB design process by simplifying complex commands into intuitive language-based interactions. By harnessing the powerful language capabilities of ChatGPT and the rich design functions of KiCad, the plugin effectively bridges the gap between complex EDA software and user-friendly interaction. Meanwhile, the new paradigm behind SmartonAI can also extend to other complex software systems, illustrating the immense potential of AI-assisted user interfaces in advancing digital interactions across various domains.
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020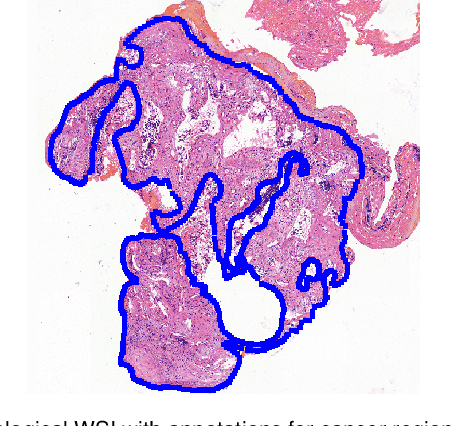
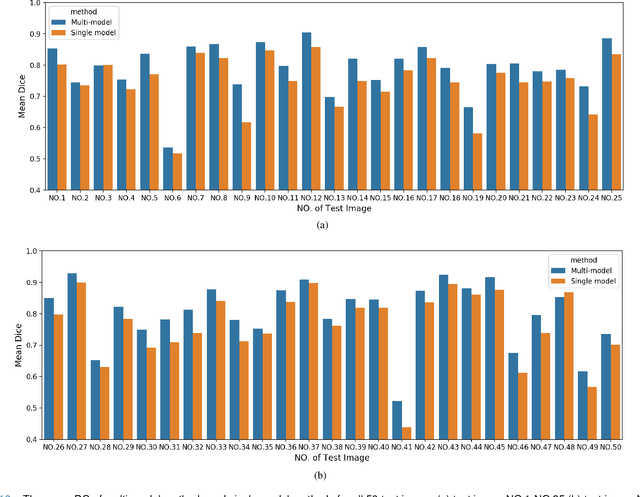
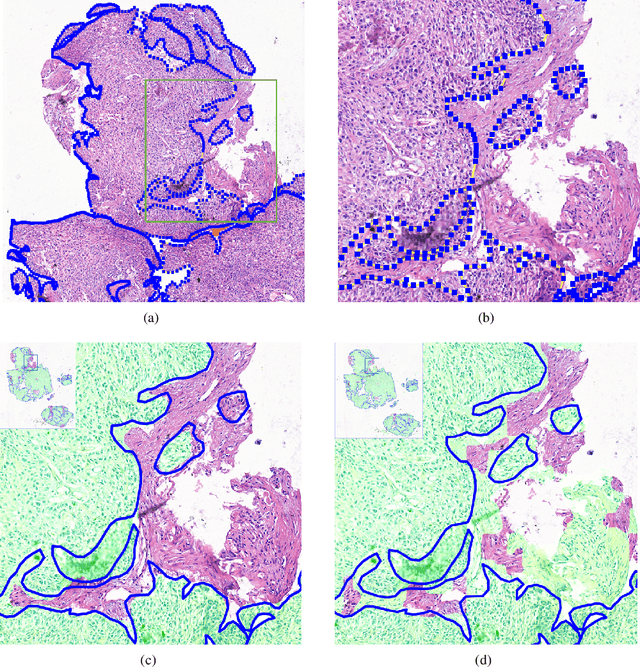
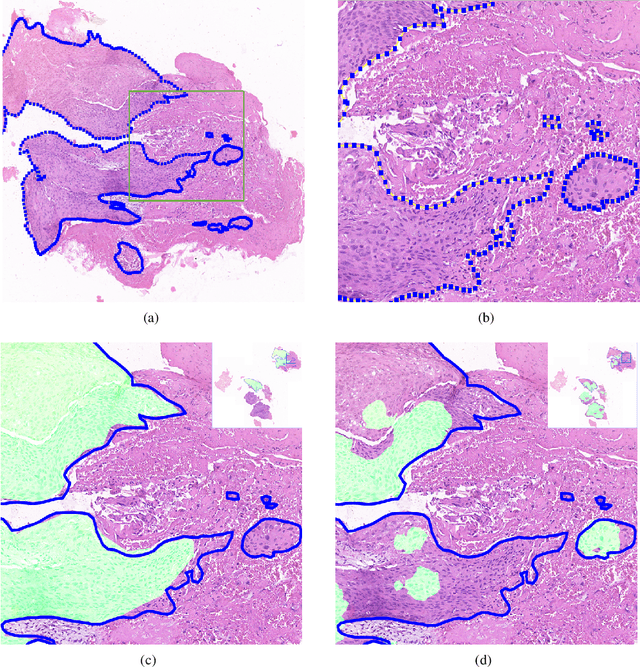
Abstract:Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge