Jianhui Zhong
High-resolution myelin-water fraction and quantitative relaxation mapping using 3D ViSTa-MR fingerprinting
Dec 21, 2023Abstract:Purpose: This study aims to develop a high-resolution whole-brain multi-parametric quantitative MRI approach for simultaneous mapping of myelin-water fraction (MWF), T1, T2, and proton-density (PD), all within a clinically feasible scan time. Methods: We developed 3D ViSTa-MRF, which combined Visualization of Short Transverse relaxation time component (ViSTa) technique with MR Fingerprinting (MRF), to achieve high-fidelity whole-brain MWF and T1/T2/PD mapping on a clinical 3T scanner. To achieve fast acquisition and memory-efficient reconstruction, the ViSTa-MRF sequence leverages an optimized 3D tiny-golden-angle-shuffling spiral-projection acquisition and joint spatial-temporal subspace reconstruction with optimized preconditioning algorithm. With the proposed ViSTa-MRF approach, high-fidelity direct MWF mapping was achieved without a need for multi-compartment fitting that could introduce bias and/or noise from additional assumptions or priors. Results: The in-vivo results demonstrate the effectiveness of the proposed acquisition and reconstruction framework to provide fast multi-parametric mapping with high SNR and good quality. The in-vivo results of 1mm- and 0.66mm-iso datasets indicate that the MWF values measured by the proposed method are consistent with standard ViSTa results that are 30x slower with lower SNR. Furthermore, we applied the proposed method to enable 5-minute whole-brain 1mm-iso assessment of MWF and T1/T2/PD mappings for infant brain development and for post-mortem brain samples. Conclusions: In this work, we have developed a 3D ViSTa-MRF technique that enables the acquisition of whole-brain MWF, quantitative T1, T2, and PD maps at 1mm and 0.66mm isotropic resolution in 5 and 15 minutes, respectively. This advancement allows for quantitative investigations of myelination changes in the brain.
Optimized multi-axis spiral projection MR fingerprinting with subspace reconstruction for rapid whole-brain high-isotropic-resolution quantitative imaging
Aug 12, 2021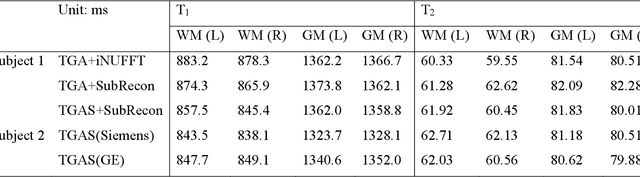
Abstract:Purpose: To improve image quality and accelerate the acquisition of 3D MRF. Methods: Building on the multi-axis spiral-projection MRF technique, a subspace reconstruction with locally low rank (LLR) constraint and a modified spiral-projection spatiotemporal encoding scheme termed tiny-golden-angle-shuffling (TGAS) were implemented for rapid whole-brain high-resolution quantitative mapping. The LLR regularization parameter and the number of subspace bases were tuned using retrospective in-vivo data and simulated examinations, respectively. B0 inhomogeneity correction using multi-frequency interpolation was incorporated into the subspace reconstruction to further improve the image quality by mitigating blurring caused by off-resonance effect. Results: The proposed MRF acquisition and reconstruction framework can produce provide high quality 1-mm isotropic whole-brain quantitative maps in a total acquisition time of 1 minute 55 seconds, with higher-quality results than ones obtained from the previous approach in 6 minutes. The comparison of quantitative results indicates that neither the subspace reconstruction nor the TGAS trajectory induce bias for T1 and T2 mapping. High quality whole-brain MRF data were also obtained at 0.66-mm isotropic resolution in 4 minutes using the proposed technique, where the increased resolution was shown to improve visualization of subtle brain structures. Conclusion: The proposed TGAS-SPI-MRF with optimized spiral-projection trajectory and subspace reconstruction can enable high-resolution quantitative mapping with faster acquisition speed.
Model-based Synthetic Data-driven Learning (MOST-DL): Application in Single-shot T2 Mapping with Severe Head Motion Using Overlapping-echo Acquisition
Jul 30, 2021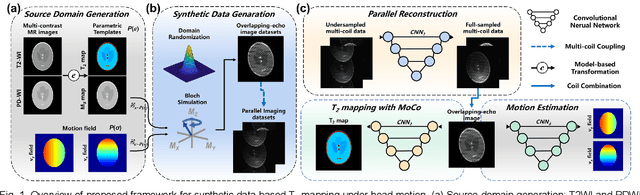
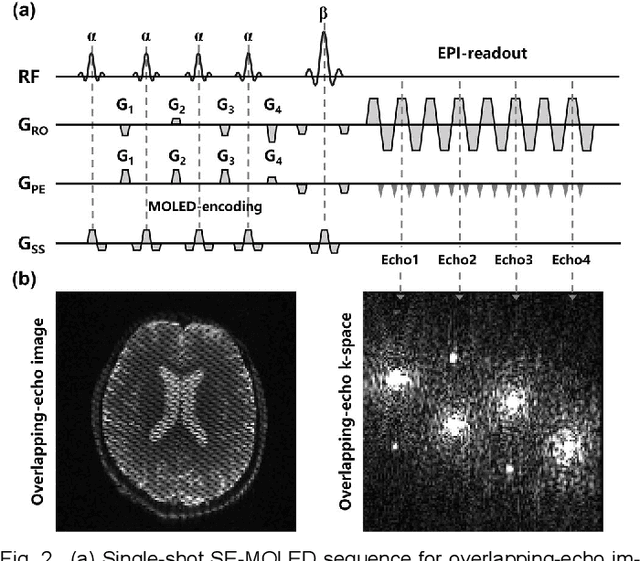
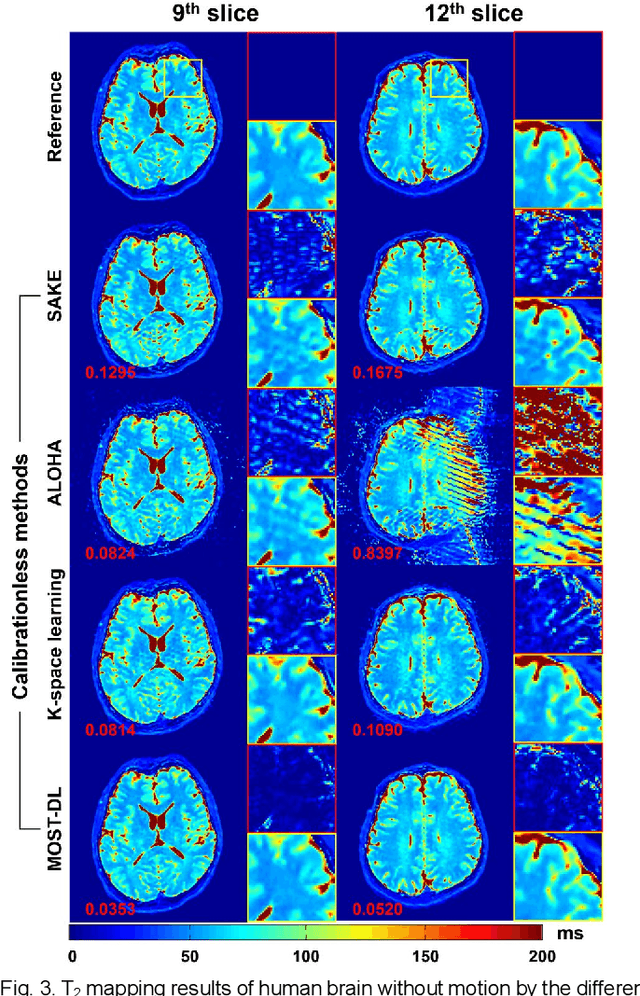
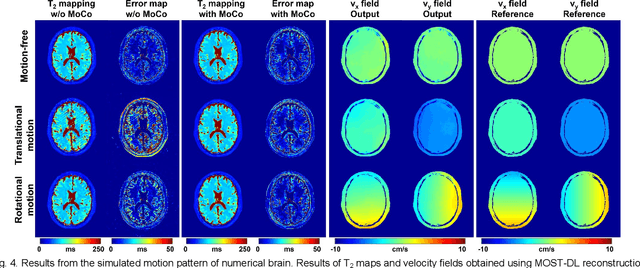
Abstract:Data-driven learning algorithm has been successfully applied to facilitate reconstruction of medical imaging. However, real-world data needed for supervised learning are typically unavailable or insufficient, especially in the field of magnetic resonance imaging (MRI). Synthetic training samples have provided a potential solution for such problem, while the challenge brought by various non-ideal situations were usually encountered especially under complex experimental conditions. In this study, a general framework, Model-based Synthetic Data-driven Learning (MOST-DL), was proposed to generate paring data for network training to achieve robust T2 mapping using overlapping-echo acquisition under severe head motion accompanied with inhomogeneous RF field. We decomposed this challenging task into parallel reconstruction and motion correction according to a forward model. The neural network was first trained in pure synthetic dataset and then evaluated with in vivo human brain. Experiments showed that MOST-DL method significantly reduces ghosting and motion artifacts in T2 maps in the presence of random and continuous subject movement. We believe that the proposed approach may open a door for solving similar problems with other MRI acquisition methods and can be extended to other areas of medical imaging.
DLpN: Single-Shell NODDI Using Deep Learner Estimated Isotropic Volume Fraction
Feb 02, 2021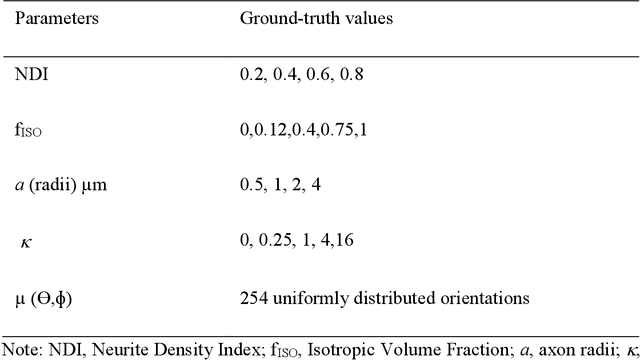
Abstract:Neurite orientation dispersion and density imaging (NODDI) enables assessment of intracellular, extracellular and free water signals from multi-shell diffusion MRI data. It is an insightful approach to characterize the brain tissue microstructure. Single-shell reconstruction for NODDI parameters has been discouraged in previous literature based on failure when fitting especially for the neurite density index (NDI). Here, we investigated the possibility to create robust NODDI parameter maps with single-shell data, using isotropic volume fraction (f_{ISO}) as prior. We made the prior estimation independent of NODDI model constraint using a dictionary based deep learning approach. First, we proposed a stochastic sparse dictionary-based network, DictNet in predicting f_{ISO} . In single-shell cases, fractional anisotropy (FA) and T2 signal without diffusion weighting ( S_0 ) were incorporated in the dictionary for f_{ISO} estimation. Then, NODDI framework was used in a prior setting to estimate the NDI and orientation dispersion index (ODI). Using both synthetic data simulation and human data collected on a 3T scanner, we compared the performance of our dictionary based deep learning prior NODDI (DLpN) with original NODDI method for both single-shell and multi-shell data. Our results suggest that DLpN derived NDI and ODI parameters for single-shell protocols are comparable with original multi-shell NODDI, and protocol with b=2000 s/mm 2 performs the best (error ~2% in white matter and ~4% in grey matter). This may allow NODDI evaluation of retrospective studies on single-shell data by additional scanning of two subjects for DictNet f_{ISO} training.
High Efficient Reconstruction of Single-shot T2 Mapping from OverLapping-Echo Detachment Planar Imaging Based on Deep Residual Network
Aug 17, 2017



Abstract:Purpose: An end-to-end deep convolutional neural network (CNN) based on deep residual network (ResNet) was proposed to efficiently reconstruct reliable T2 mapping from single-shot OverLapping-Echo Detachment (OLED) planar imaging. Methods: The training dataset was obtained from simulations carried out on SPROM software developed by our group. The relationship between the original OLED image containing two echo signals and the corresponded T2 mapping was learned by ResNet training. After the ResNet was trained, it was applied to reconstruct the T2 mapping from simulation and in vivo human brain data. Results: Though the ResNet was trained entirely on simulated data, the trained network was generalized well to real human brain data. The results from simulation and in vivo human brain experiments show that the proposed method significantly outperformed the echo-detachment-based method. Reliable T2 mapping was achieved within tens of milliseconds after the network had been trained while the echo-detachment-based OLED reconstruction method took minutes. Conclusion: The proposed method will greatly facilitate real-time dynamic and quantitative MR imaging via OLED sequence, and ResNet has the potential to reconstruct images from complex MRI sequence efficiently.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge