DLpN: Single-Shell NODDI Using Deep Learner Estimated Isotropic Volume Fraction
Paper and Code
Feb 02, 2021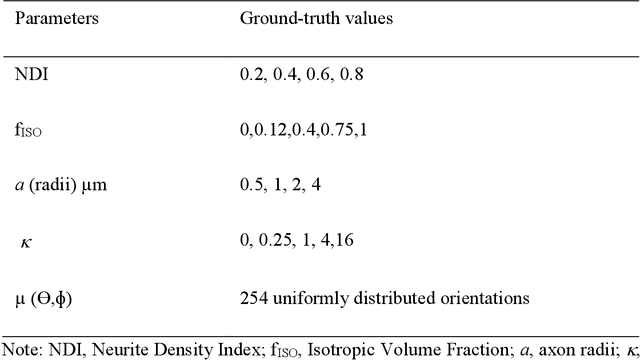
Neurite orientation dispersion and density imaging (NODDI) enables assessment of intracellular, extracellular and free water signals from multi-shell diffusion MRI data. It is an insightful approach to characterize the brain tissue microstructure. Single-shell reconstruction for NODDI parameters has been discouraged in previous literature based on failure when fitting especially for the neurite density index (NDI). Here, we investigated the possibility to create robust NODDI parameter maps with single-shell data, using isotropic volume fraction (f_{ISO}) as prior. We made the prior estimation independent of NODDI model constraint using a dictionary based deep learning approach. First, we proposed a stochastic sparse dictionary-based network, DictNet in predicting f_{ISO} . In single-shell cases, fractional anisotropy (FA) and T2 signal without diffusion weighting ( S_0 ) were incorporated in the dictionary for f_{ISO} estimation. Then, NODDI framework was used in a prior setting to estimate the NDI and orientation dispersion index (ODI). Using both synthetic data simulation and human data collected on a 3T scanner, we compared the performance of our dictionary based deep learning prior NODDI (DLpN) with original NODDI method for both single-shell and multi-shell data. Our results suggest that DLpN derived NDI and ODI parameters for single-shell protocols are comparable with original multi-shell NODDI, and protocol with b=2000 s/mm 2 performs the best (error ~2% in white matter and ~4% in grey matter). This may allow NODDI evaluation of retrospective studies on single-shell data by additional scanning of two subjects for DictNet f_{ISO} training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge