Jake Sunshine
Scaling Wearable Foundation Models
Oct 17, 2024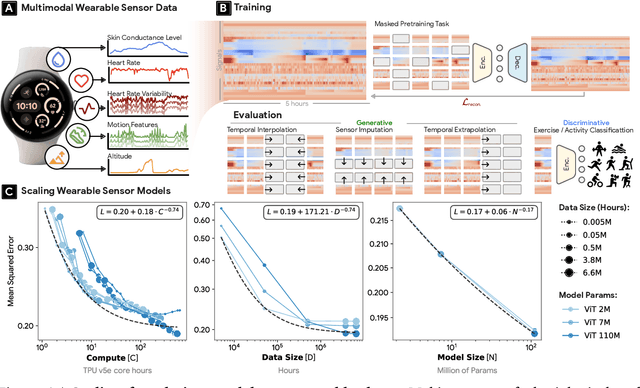
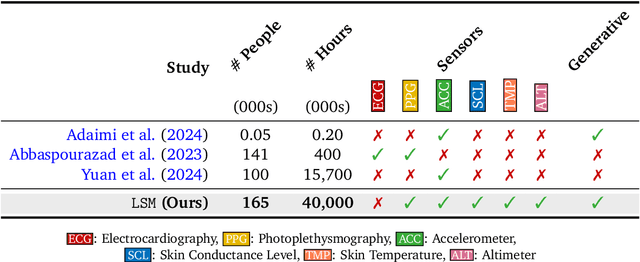


Abstract:Wearable sensors have become ubiquitous thanks to a variety of health tracking features. The resulting continuous and longitudinal measurements from everyday life generate large volumes of data; however, making sense of these observations for scientific and actionable insights is non-trivial. Inspired by the empirical success of generative modeling, where large neural networks learn powerful representations from vast amounts of text, image, video, or audio data, we investigate the scaling properties of sensor foundation models across compute, data, and model size. Using a dataset of up to 40 million hours of in-situ heart rate, heart rate variability, electrodermal activity, accelerometer, skin temperature, and altimeter per-minute data from over 165,000 people, we create LSM, a multimodal foundation model built on the largest wearable-signals dataset with the most extensive range of sensor modalities to date. Our results establish the scaling laws of LSM for tasks such as imputation, interpolation and extrapolation, both across time and sensor modalities. Moreover, we highlight how LSM enables sample-efficient downstream learning for tasks like exercise and activity recognition.
Transforming Wearable Data into Health Insights using Large Language Model Agents
Jun 11, 2024



Abstract:Despite the proliferation of wearable health trackers and the importance of sleep and exercise to health, deriving actionable personalized insights from wearable data remains a challenge because doing so requires non-trivial open-ended analysis of these data. The recent rise of large language model (LLM) agents, which can use tools to reason about and interact with the world, presents a promising opportunity to enable such personalized analysis at scale. Yet, the application of LLM agents in analyzing personal health is still largely untapped. In this paper, we introduce the Personal Health Insights Agent (PHIA), an agent system that leverages state-of-the-art code generation and information retrieval tools to analyze and interpret behavioral health data from wearables. We curate two benchmark question-answering datasets of over 4000 health insights questions. Based on 650 hours of human and expert evaluation we find that PHIA can accurately address over 84% of factual numerical questions and more than 83% of crowd-sourced open-ended questions. This work has implications for advancing behavioral health across the population, potentially enabling individuals to interpret their own wearable data, and paving the way for a new era of accessible, personalized wellness regimens that are informed by data-driven insights.
Towards Accurate Differential Diagnosis with Large Language Models
Nov 30, 2023



Abstract:An accurate differential diagnosis (DDx) is a cornerstone of medical care, often reached through an iterative process of interpretation that combines clinical history, physical examination, investigations and procedures. Interactive interfaces powered by Large Language Models (LLMs) present new opportunities to both assist and automate aspects of this process. In this study, we introduce an LLM optimized for diagnostic reasoning, and evaluate its ability to generate a DDx alone or as an aid to clinicians. 20 clinicians evaluated 302 challenging, real-world medical cases sourced from the New England Journal of Medicine (NEJM) case reports. Each case report was read by two clinicians, who were randomized to one of two assistive conditions: either assistance from search engines and standard medical resources, or LLM assistance in addition to these tools. All clinicians provided a baseline, unassisted DDx prior to using the respective assistive tools. Our LLM for DDx exhibited standalone performance that exceeded that of unassisted clinicians (top-10 accuracy 59.1% vs 33.6%, [p = 0.04]). Comparing the two assisted study arms, the DDx quality score was higher for clinicians assisted by our LLM (top-10 accuracy 51.7%) compared to clinicians without its assistance (36.1%) (McNemar's Test: 45.7, p < 0.01) and clinicians with search (44.4%) (4.75, p = 0.03). Further, clinicians assisted by our LLM arrived at more comprehensive differential lists than those without its assistance. Our study suggests that our LLM for DDx has potential to improve clinicians' diagnostic reasoning and accuracy in challenging cases, meriting further real-world evaluation for its ability to empower physicians and widen patients' access to specialist-level expertise.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge