Ismael Boureima
Theoretical Division, Los Alamos National Laboratory, Los Alamos, USA
Enhancing Cross-Language Code Translation via Task-Specific Embedding Alignment in Retrieval-Augmented Generation
Dec 06, 2024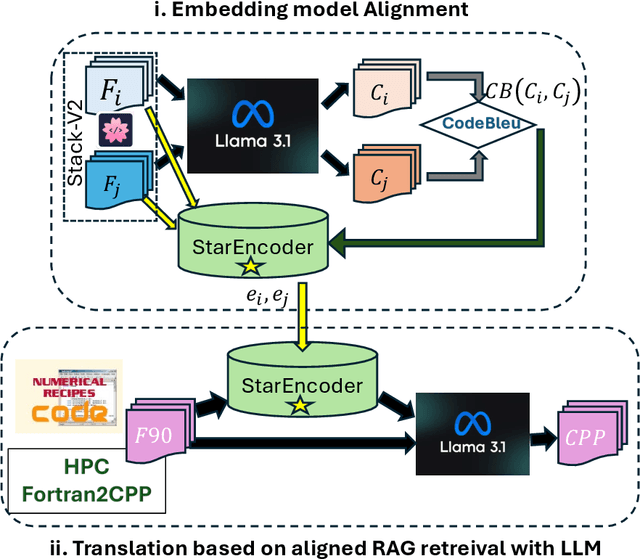



Abstract:We introduce a novel method to enhance cross-language code translation from Fortran to C++ by integrating task-specific embedding alignment into a Retrieval-Augmented Generation (RAG) framework. Unlike conventional retrieval approaches that utilize generic embeddings agnostic to the downstream task, our strategy aligns the retrieval model directly with the objective of maximizing translation quality, as quantified by the CodeBLEU metric. This alignment ensures that the embeddings are semantically and syntactically meaningful for the specific code translation task. Our methodology involves constructing a dataset of 25,000 Fortran code snippets sourced from Stack-V2 dataset and generating their corresponding C++ translations using the LLaMA 3.1-8B language model. We compute pairwise CodeBLEU scores between the generated translations and ground truth examples to capture fine-grained similarities. These scores serve as supervision signals in a contrastive learning framework, where we optimize the embedding model to retrieve Fortran-C++ pairs that are most beneficial for improving the language model's translation performance. By integrating these CodeBLEU-optimized embeddings into the RAG framework, our approach significantly enhances both retrieval accuracy and code generation quality over methods employing generic embeddings. On the HPC Fortran2C++ dataset, our method elevates the average CodeBLEU score from 0.64 to 0.73, achieving a 14% relative improvement. On the Numerical Recipes dataset, we observe an increase from 0.52 to 0.60, marking a 15% relative improvement. Importantly, these gains are realized without any fine-tuning of the language model, underscoring the efficiency and practicality of our approach.
HEAL: Hierarchical Embedding Alignment Loss for Improved Retrieval and Representation Learning
Dec 05, 2024Abstract:Retrieval-Augmented Generation (RAG) enhances Large Language Models (LLMs) by integrating external document retrieval to provide domain-specific or up-to-date knowledge. The effectiveness of RAG depends on the relevance of retrieved documents, which is influenced by the semantic alignment of embeddings with the domain's specialized content. Although full fine-tuning can align language models to specific domains, it is computationally intensive and demands substantial data. This paper introduces Hierarchical Embedding Alignment Loss (HEAL), a novel method that leverages hierarchical fuzzy clustering with matrix factorization within contrastive learning to efficiently align LLM embeddings with domain-specific content. HEAL computes level/depth-wise contrastive losses and incorporates hierarchical penalties to align embeddings with the underlying relationships in label hierarchies. This approach enhances retrieval relevance and document classification, effectively reducing hallucinations in LLM outputs. In our experiments, we benchmark and evaluate HEAL across diverse domains, including Healthcare, Material Science, Cyber-security, and Applied Maths.
Tensor Train Low-rank Approximation (TT-LoRA): Democratizing AI with Accelerated LLMs
Aug 02, 2024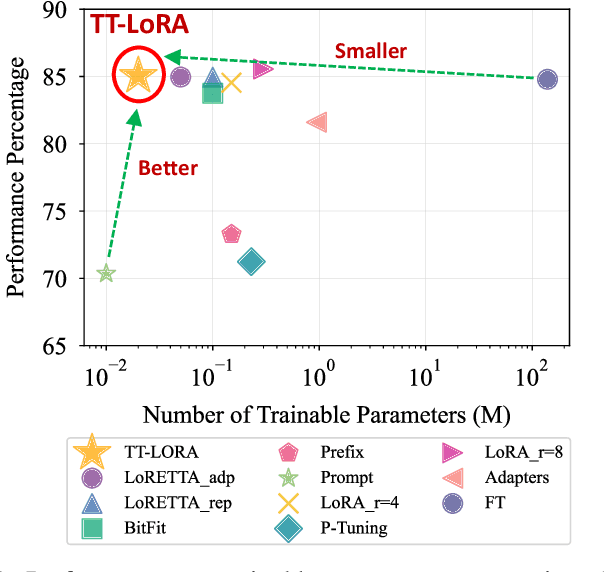
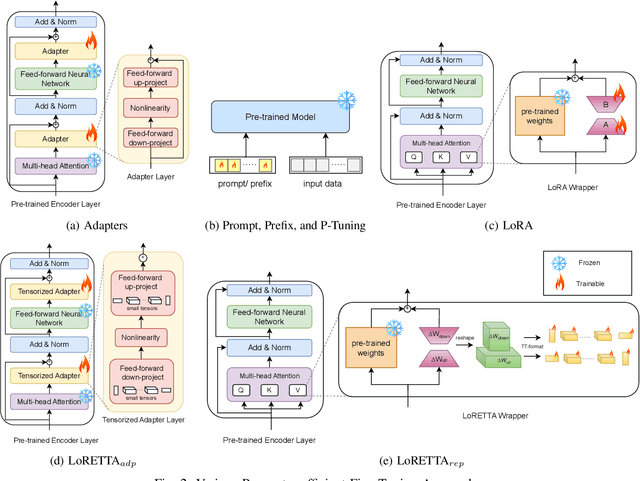
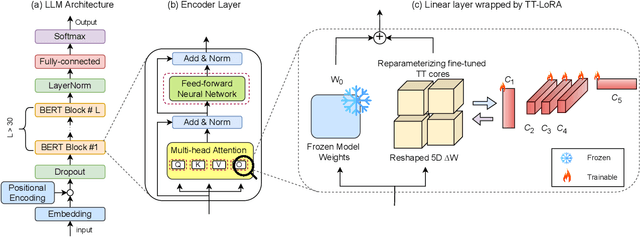
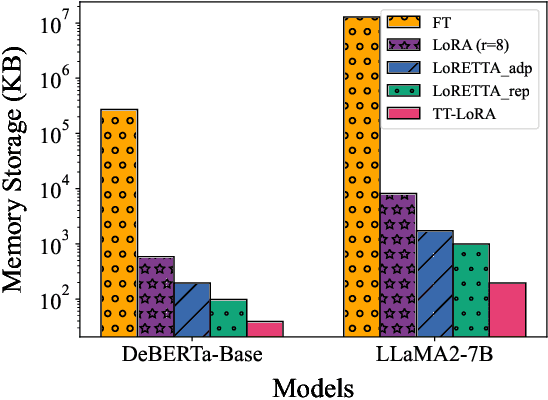
Abstract:In recent years, Large Language Models (LLMs) have demonstrated remarkable capabilities across a wide range of natural language processing (NLP) tasks, such as question-answering, sentiment analysis, text summarization, and machine translation. However, the ever-growing complexity of LLMs demands immense computational resources, hindering the broader research and application of these models. To address this, various parameter-efficient fine-tuning strategies, such as Low-Rank Approximation (LoRA) and Adapters, have been developed. Despite their potential, these methods often face limitations in compressibility. Specifically, LoRA struggles to scale effectively with the increasing number of trainable parameters in modern large scale LLMs. Additionally, Low-Rank Economic Tensor-Train Adaptation (LoRETTA), which utilizes tensor train decomposition, has not yet achieved the level of compression necessary for fine-tuning very large scale models with limited resources. This paper introduces Tensor Train Low-Rank Approximation (TT-LoRA), a novel parameter-efficient fine-tuning (PEFT) approach that extends LoRETTA with optimized tensor train (TT) decomposition integration. By eliminating Adapters and traditional LoRA-based structures, TT-LoRA achieves greater model compression without compromising downstream task performance, along with reduced inference latency and computational overhead. We conduct an exhaustive parameter search to establish benchmarks that highlight the trade-off between model compression and performance. Our results demonstrate significant compression of LLMs while maintaining comparable performance to larger models, facilitating their deployment on resource-constraint platforms.
Binary Bleed: Fast Distributed and Parallel Method for Automatic Model Selection
Jul 26, 2024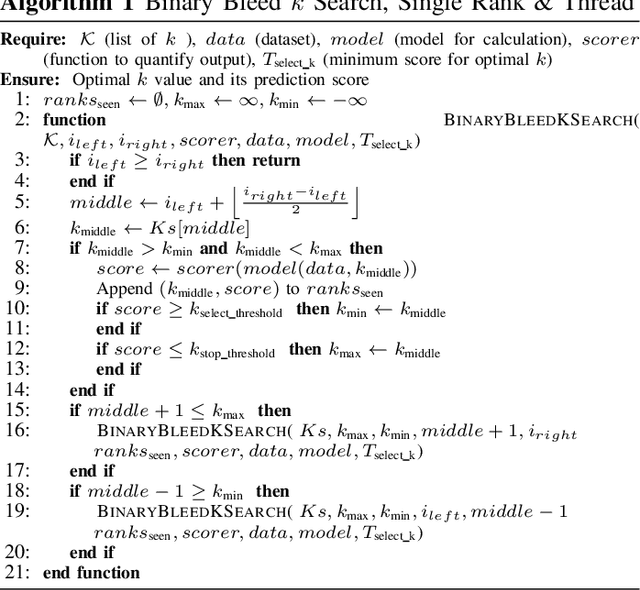

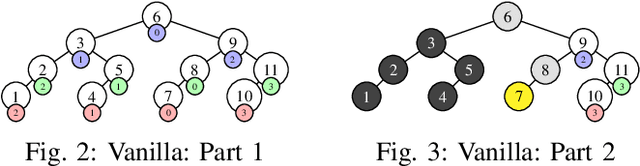
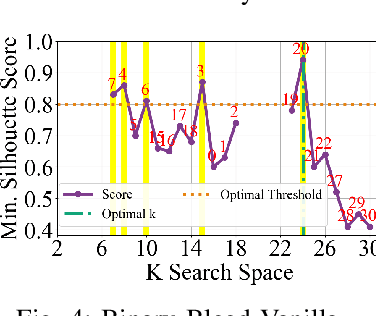
Abstract:In several Machine Learning (ML) clustering and dimensionality reduction approaches, such as non-negative matrix factorization (NMF), RESCAL, and K-Means clustering, users must select a hyper-parameter k to define the number of clusters or components that yield an ideal separation of samples or clean clusters. This selection, while difficult, is crucial to avoid overfitting or underfitting the data. Several ML applications use scoring methods (e.g., Silhouette and Davies Boulding scores) to evaluate the cluster pattern stability for a specific k. The score is calculated for different trials over a range of k, and the ideal k is heuristically selected as the value before the model starts overfitting, indicated by a drop or increase in the score resembling an elbow curve plot. While the grid-search method can be used to accurately find a good k value, visiting a range of k can become time-consuming and computationally resource-intensive. In this paper, we introduce the Binary Bleed method based on binary search, which significantly reduces the k search space for these grid-search ML algorithms by truncating the target k values from the search space using a heuristic with thresholding over the scores. Binary Bleed is designed to work with single-node serial, single-node multi-processing, and distributed computing resources. In our experiments, we demonstrate the reduced search space gain over a naive sequential search of the ideal k and the accuracy of the Binary Bleed in identifying the correct k for NMFk, K-Means pyDNMFk, and pyDRESCALk with Silhouette and Davies Boulding scores. We make our implementation of Binary Bleed for the NMF algorithm available on GitHub.
Distributed Out-of-Memory NMF of Dense and Sparse Data on CPU/GPU Architectures with Automatic Model Selection for Exascale Data
Feb 19, 2022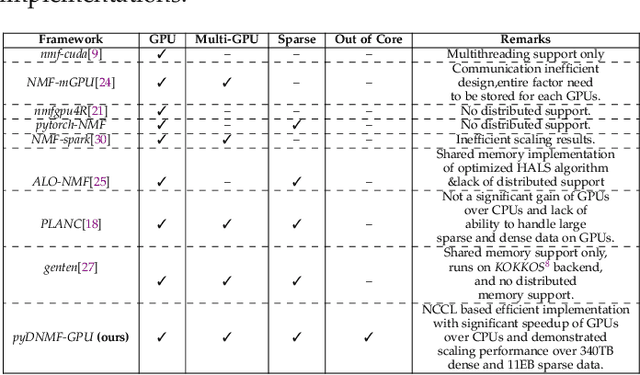
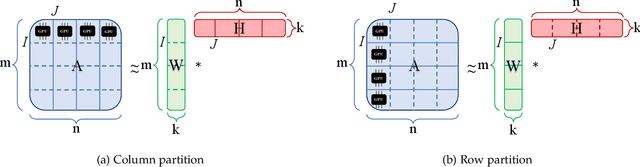
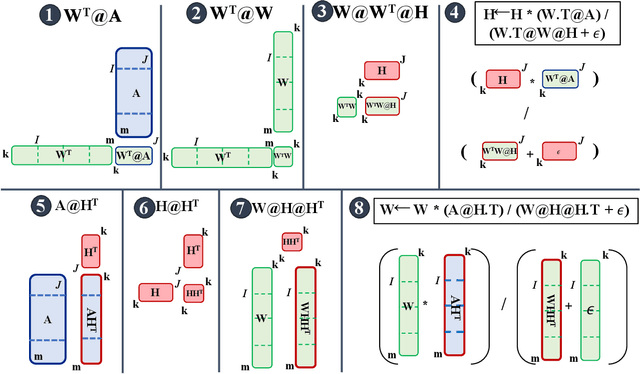
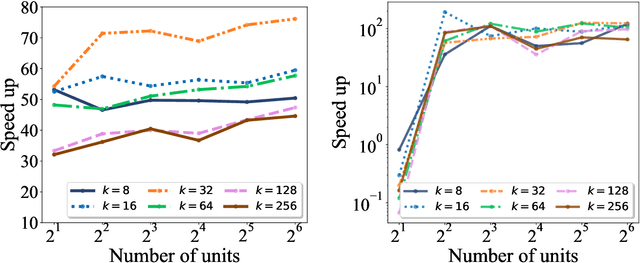
Abstract:The need for efficient and scalable big-data analytics methods is more essential than ever due to the exploding size and complexity of globally emerging datasets. Nonnegative Matrix Factorization (NMF) is a well-known explainable unsupervised learning method for dimensionality reduction, latent feature extraction, blind source separation, data mining, and machine learning. In this paper, we introduce a new distributed out-of-memory NMF method, named pyDNMF-GPU, designed for modern heterogeneous CPU/GPU architectures that is capable of factoring exascale-sized dense and sparse matrices. Our method reduces the latency associated with local data transfer between the GPU and host using CUDA streams, and reduces the latency associated with collective communications (both intra-node and inter-node) via NCCL primitives. In addition, sparse and dense matrix multiplications are significantly accelerated with GPU cores, resulting in good scalability. We set new benchmarks for the size of the data being analyzed: in experiments, we measure up to 76x improvement on a single GPU over running on a single 18 core CPU and we show good weak scaling on up to 4096 multi-GPU cluster nodes with approximately 25,000 GPUs, when decomposing a dense 340 Terabyte-size matrix and a 11 Exabyte-size sparse matrix of density 10e-6. Finally, we integrate our method with an automatic model selection method. With this integration, we introduce a new tool that is capable of analyzing, compressing, and discovering explainable latent structures in extremely large sparse and dense data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge