Hyun Jung
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Speak or Chat with Me: End-to-End Spoken Language Understanding System with Flexible Inputs
Apr 07, 2021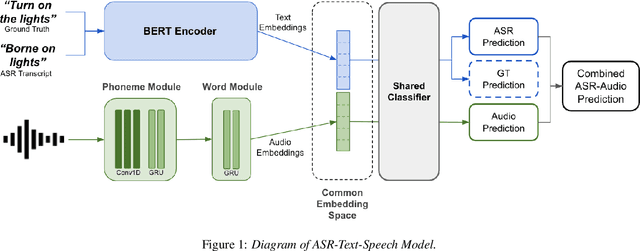
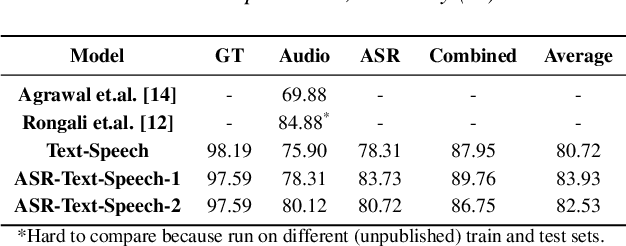
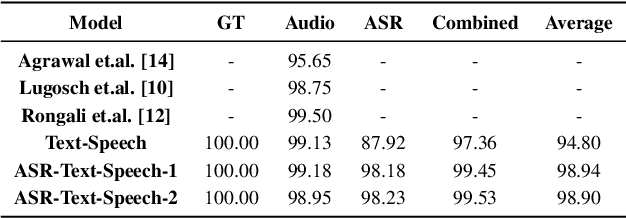
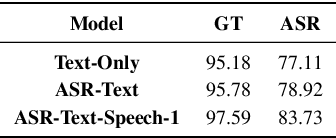
Abstract:A major focus of recent research in spoken language understanding (SLU) has been on the end-to-end approach where a single model can predict intents directly from speech inputs without intermediate transcripts. However, this approach presents some challenges. First, since speech can be considered as personally identifiable information, in some cases only automatic speech recognition (ASR) transcripts are accessible. Second, intent-labeled speech data is scarce. To address the first challenge, we propose a novel system that can predict intents from flexible types of inputs: speech, ASR transcripts, or both. We demonstrate strong performance for either modality separately, and when both speech and ASR transcripts are available, through system combination, we achieve better results than using a single input modality. To address the second challenge, we leverage a semantically robust pre-trained BERT model and adopt a cross-modal system that co-trains text embeddings and acoustic embeddings in a shared latent space. We further enhance this system by utilizing an acoustic module pre-trained on LibriSpeech and domain-adapting the text module on our target datasets. Our experiments show significant advantages for these pre-training and fine-tuning strategies, resulting in a system that achieves competitive intent-classification performance on Snips SLU and Fluent Speech Commands datasets.
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020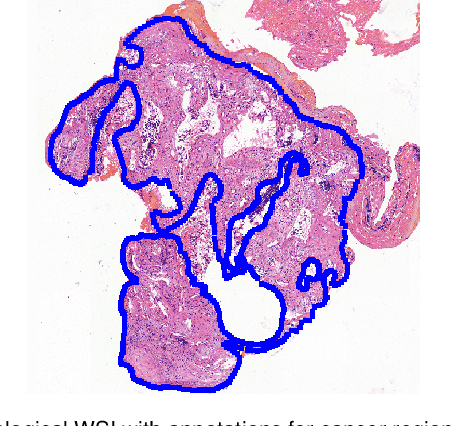
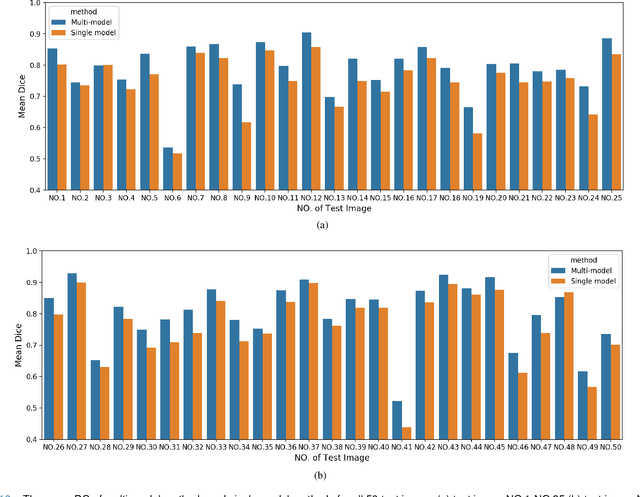
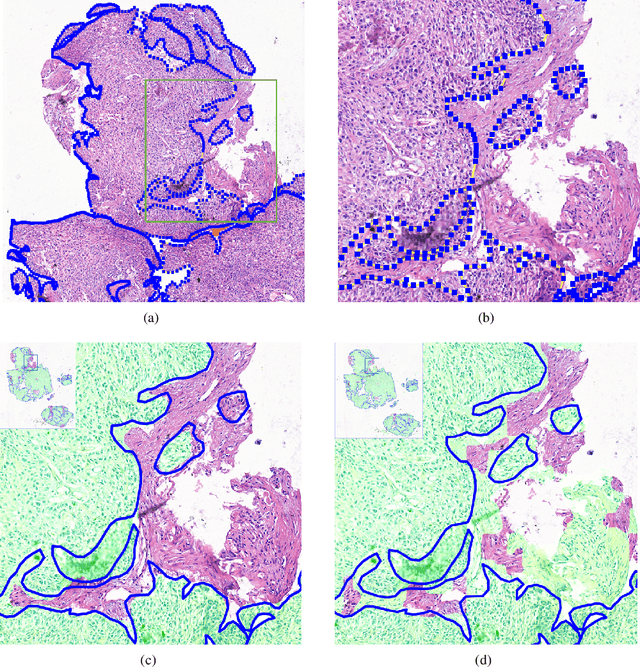
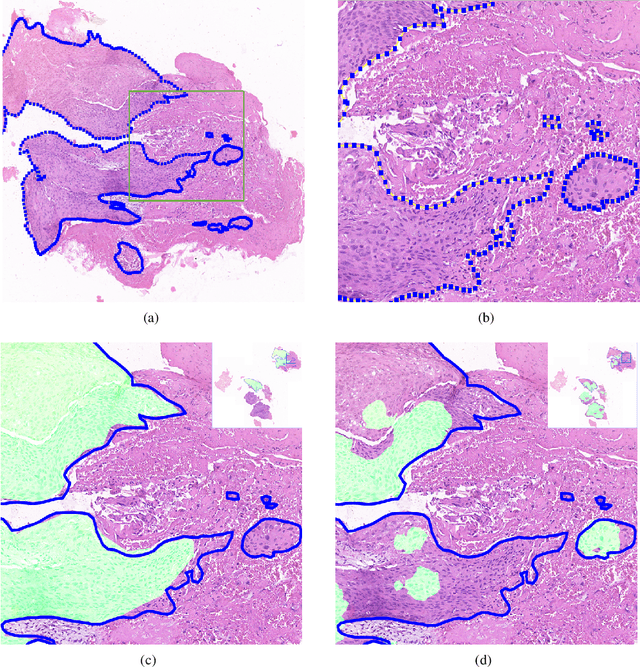
Abstract:Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge