Guorong Wu
GeoDynamics: A Geometric State-Space Neural Network for Understanding Brain Dynamics on Riemannian Manifolds
Jan 20, 2026Abstract:State-space models (SSMs) have become a cornerstone for unraveling brain dynamics, revealing how latent neural states evolve over time and give rise to observed signals. By combining the flexibility of deep learning with the principled dynamical structure of SSMs, recent studies have achieved powerful fits to functional neuroimaging data. However, most existing approaches still view the brain as a set of loosely connected regions or impose oversimplified network priors, falling short of a truly holistic and self-organized dynamical system perspective. Brain functional connectivity (FC) at each time point naturally forms a symmetric positive definite (SPD) matrix, which resides on a curved Riemannian manifold rather than in Euclidean space. Capturing the trajectories of these SPD matrices is key to understanding how coordinated networks support cognition and behavior. To this end, we introduce GeoDynamics, a geometric state-space neural network that tracks latent brain-state trajectories directly on the high-dimensional SPD manifold. GeoDynamics embeds each connectivity matrix into a manifold-aware recurrent framework, learning smooth and geometry-respecting transitions that reveal task-driven state changes and early markers of Alzheimer's disease, Parkinson's disease, and autism. Beyond neuroscience, we validate GeoDynamics on human action recognition benchmarks (UTKinect, Florence, HDM05), demonstrating its scalability and robustness in modeling complex spatiotemporal dynamics across diverse domains.
De-Individualizing fMRI Signals via Mahalanobis Whitening and Bures Geometry
Nov 10, 2025Abstract:Functional connectivity has been widely investigated to understand brain disease in clinical studies and imaging-based neuroscience, and analyzing changes in functional connectivity has proven to be valuable for understanding and computationally evaluating the effects on brain function caused by diseases or experimental stimuli. By using Mahalanobis data whitening prior to the use of dimensionality reduction algorithms, we are able to distill meaningful information from fMRI signals about subjects and the experimental stimuli used to prompt them. Furthermore, we offer an interpretation of Mahalanobis whitening as a two-stage de-individualization of data which is motivated by similarity as captured by the Bures distance, which is connected to quantum mechanics. These methods have potential to aid discoveries about the mechanisms that link brain function with cognition and behavior and may improve the accuracy and consistency of Alzheimer's diagnosis, especially in the preclinical stage of disease progression.
OCL: Ordinal Contrastive Learning for Imputating Features with Progressive Labels
Mar 03, 2025Abstract:Accurately discriminating progressive stages of Alzheimer's Disease (AD) is crucial for early diagnosis and prevention. It often involves multiple imaging modalities to understand the complex pathology of AD, however, acquiring a complete set of images is challenging due to high cost and burden for subjects. In the end, missing data become inevitable which lead to limited sample-size and decrease in precision in downstream analyses. To tackle this challenge, we introduce a holistic imaging feature imputation method that enables to leverage diverse imaging features while retaining all subjects. The proposed method comprises two networks: 1) An encoder to extract modality-independent embeddings and 2) A decoder to reconstruct the original measures conditioned on their imaging modalities. The encoder includes a novel {\em ordinal contrastive loss}, which aligns samples in the embedding space according to the progression of AD. We also maximize modality-wise coherence of embeddings within each subject, in conjunction with domain adversarial training algorithms, to further enhance alignment between different imaging modalities. The proposed method promotes our holistic imaging feature imputation across various modalities in the shared embedding space. In the experiments, we show that our networks deliver favorable results for statistical analysis and classification against imputation baselines with Alzheimer's Disease Neuroimaging Initiative (ADNI) study.
Modality-Agnostic Style Transfer for Holistic Feature Imputation
Mar 03, 2025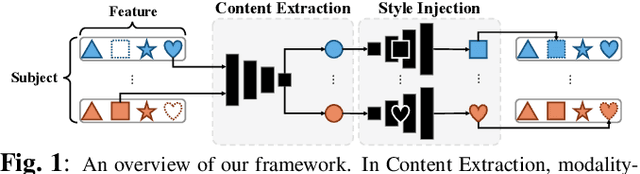
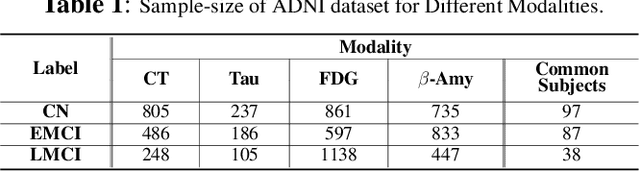
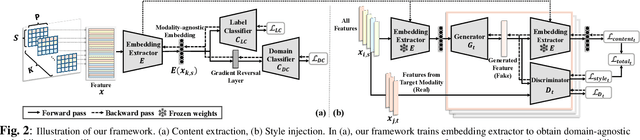
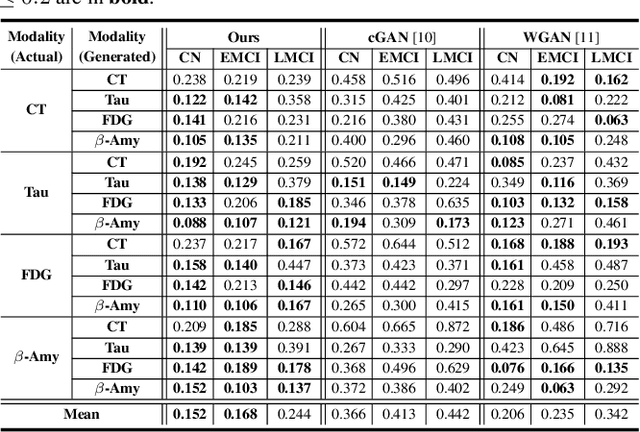
Abstract:Characterizing a preclinical stage of Alzheimer's Disease (AD) via single imaging is difficult as its early symptoms are quite subtle. Therefore, many neuroimaging studies are curated with various imaging modalities, e.g., MRI and PET, however, it is often challenging to acquire all of them from all subjects and missing data become inevitable. In this regards, in this paper, we propose a framework that generates unobserved imaging measures for specific subjects using their existing measures, thereby reducing the need for additional examinations. Our framework transfers modality-specific style while preserving AD-specific content. This is done by domain adversarial training that preserves modality-agnostic but AD-specific information, while a generative adversarial network adds an indistinguishable modality-specific style. Our proposed framework is evaluated on the Alzheimer's Disease Neuroimaging Initiative (ADNI) study and compared with other imputation methods in terms of generated data quality. Small average Cohen's $d$ $< 0.19$ between our generated measures and real ones suggests that the synthetic data are practically usable regardless of their modality type.
Learning Covariance-Based Multi-Scale Representation of Neuroimaging Measures for Alzheimer Classification
Mar 03, 2025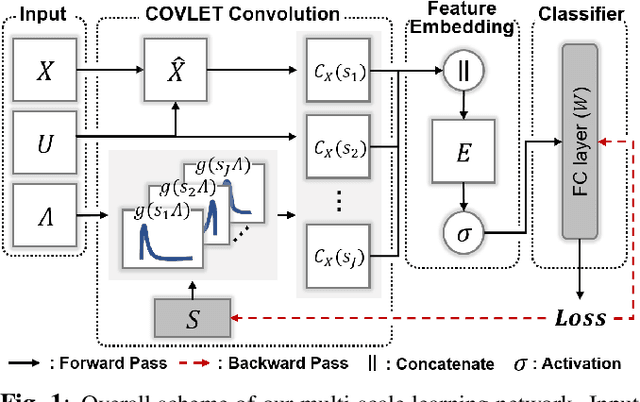
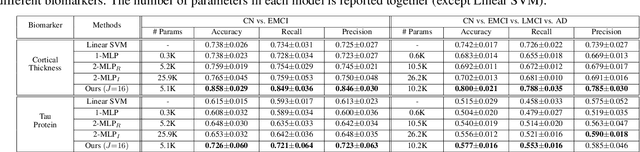
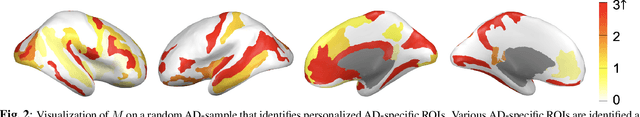
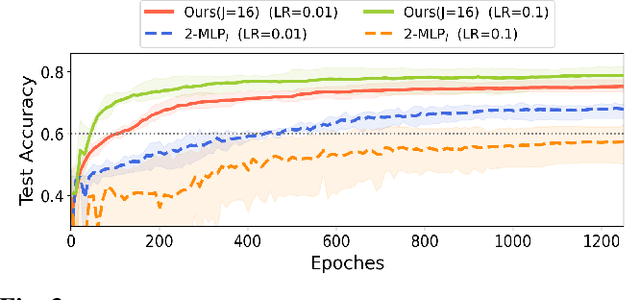
Abstract:Stacking excessive layers in DNN results in highly underdetermined system when training samples are limited, which is very common in medical applications. In this regard, we present a framework capable of deriving an efficient high-dimensional space with reasonable increase in model size. This is done by utilizing a transform (i.e., convolution) that leverages scale-space theory with covariance structure. The overall model trains on this transform together with a downstream classifier (i.e., Fully Connected layer) to capture the optimal multi-scale representation of the original data which corresponds to task-specific components in a dual space. Experiments on neuroimaging measures from Alzheimer's Disease Neuroimaging Initiative (ADNI) study show that our model performs better and converges faster than conventional models even when the model size is significantly reduced. The trained model is made interpretable using gradient information over the multi-scale transform to delineate personalized AD-specific regions in the brain.
BrainMAP: Learning Multiple Activation Pathways in Brain Networks
Dec 23, 2024Abstract:Functional Magnetic Resonance Image (fMRI) is commonly employed to study human brain activity, since it offers insight into the relationship between functional fluctuations and human behavior. To enhance analysis and comprehension of brain activity, Graph Neural Networks (GNNs) have been widely applied to the analysis of functional connectivities (FC) derived from fMRI data, due to their ability to capture the synergistic interactions among brain regions. However, in the human brain, performing complex tasks typically involves the activation of certain pathways, which could be represented as paths across graphs. As such, conventional GNNs struggle to learn from these pathways due to the long-range dependencies of multiple pathways. To address these challenges, we introduce a novel framework BrainMAP to learn Multiple Activation Pathways in Brain networks. BrainMAP leverages sequential models to identify long-range correlations among sequentialized brain regions and incorporates an aggregation module based on Mixture of Experts (MoE) to learn from multiple pathways. Our comprehensive experiments highlight BrainMAP's superior performance. Furthermore, our framework enables explanatory analyses of crucial brain regions involved in tasks. Our code is provided at https://github.com/LzyFischer/Graph-Mamba.
NeuroPath: A Neural Pathway Transformer for Joining the Dots of Human Connectomes
Sep 26, 2024
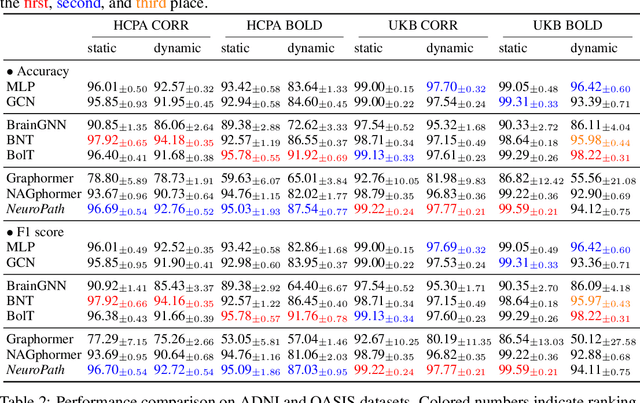

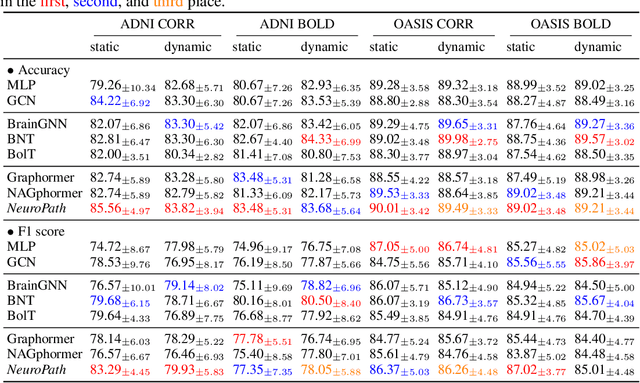
Abstract:Although modern imaging technologies allow us to study connectivity between two distinct brain regions in-vivo, an in-depth understanding of how anatomical structure supports brain function and how spontaneous functional fluctuations emerge remarkable cognition is still elusive. Meanwhile, tremendous efforts have been made in the realm of machine learning to establish the nonlinear mapping between neuroimaging data and phenotypic traits. However, the absence of neuroscience insight in the current approaches poses significant challenges in understanding cognitive behavior from transient neural activities. To address this challenge, we put the spotlight on the coupling mechanism of structural connectivity (SC) and functional connectivity (FC) by formulating such network neuroscience question into an expressive graph representation learning problem for high-order topology. Specifically, we introduce the concept of topological detour to characterize how a ubiquitous instance of FC (direct link) is supported by neural pathways (detour) physically wired by SC, which forms a cyclic loop interacted by brain structure and function. In the clich\'e of machine learning, the multi-hop detour pathway underlying SC-FC coupling allows us to devise a novel multi-head self-attention mechanism within Transformer to capture multi-modal feature representation from paired graphs of SC and FC. Taken together, we propose a biological-inspired deep model, coined as NeuroPath, to find putative connectomic feature representations from the unprecedented amount of neuroimages, which can be plugged into various downstream applications such as task recognition and disease diagnosis. We have evaluated NeuroPath on large-scale public datasets including HCP and UK Biobank under supervised and zero-shot learning, where the state-of-the-art performance by our NeuroPath indicates great potential in network neuroscience.
Machine Learning on Dynamic Functional Connectivity: Promise, Pitfalls, and Interpretations
Sep 17, 2024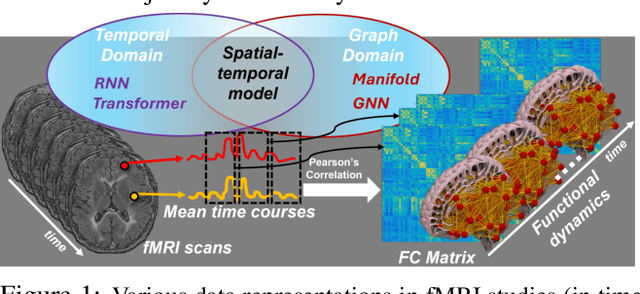
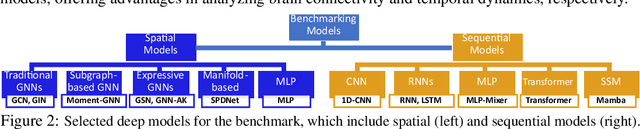
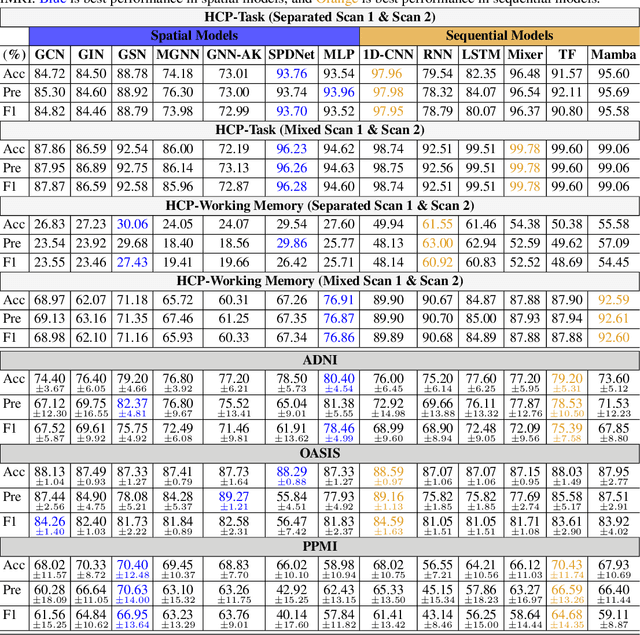
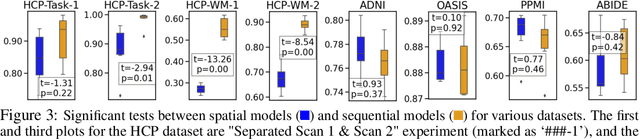
Abstract:An unprecedented amount of existing functional Magnetic Resonance Imaging (fMRI) data provides a new opportunity to understand the relationship between functional fluctuation and human cognition/behavior using a data-driven approach. To that end, tremendous efforts have been made in machine learning to predict cognitive states from evolving volumetric images of blood-oxygen-level-dependent (BOLD) signals. Due to the complex nature of brain function, however, the evaluation on learning performance and discoveries are not often consistent across current state-of-the-arts (SOTA). By capitalizing on large-scale existing neuroimaging data (34,887 data samples from six public databases), we seek to establish a well-founded empirical guideline for designing deep models for functional neuroimages by linking the methodology underpinning with knowledge from the neuroscience domain. Specifically, we put the spotlight on (1) What is the current SOTA performance in cognitive task recognition and disease diagnosis using fMRI? (2) What are the limitations of current deep models? and (3) What is the general guideline for selecting the suitable machine learning backbone for new neuroimaging applications? We have conducted a comprehensive evaluation and statistical analysis, in various settings, to answer the above outstanding questions.
Message Detouring: A Simple Yet Effective Cycle Representation for Expressive Graph Learning
Feb 12, 2024



Abstract:Graph learning is crucial in the fields of bioinformatics, social networks, and chemicals. Although high-order graphlets, such as cycles, are critical to achieving an informative graph representation for node classification, edge prediction, and graph recognition, modeling high-order topological characteristics poses significant computational challenges, restricting its widespread applications in machine learning. To address this limitation, we introduce the concept of \textit{message detouring} to hierarchically characterize cycle representation throughout the entire graph, which capitalizes on the contrast between the shortest and longest pathways within a range of local topologies associated with each graph node. The topological feature representations derived from our message detouring landscape demonstrate comparable expressive power to high-order \textit{Weisfeiler-Lehman} (WL) tests but much less computational demands. In addition to the integration with graph kernel and message passing neural networks, we present a novel message detouring neural network, which uses Transformer backbone to integrate cycle representations across nodes and edges. Aside from theoretical results, experimental results on expressiveness, graph classification, and node classification show message detouring can significantly outperform current counterpart approaches on various benchmark datasets.
Learning to Approximate Adaptive Kernel Convolution on Graphs
Jan 22, 2024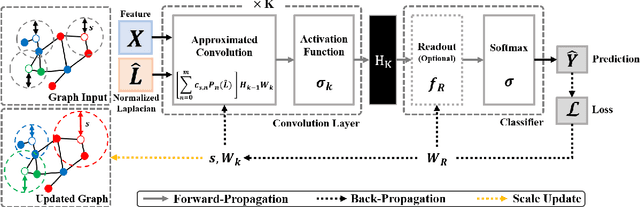
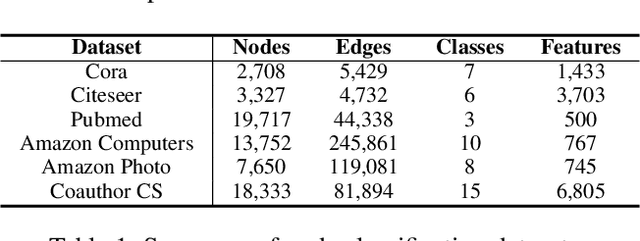
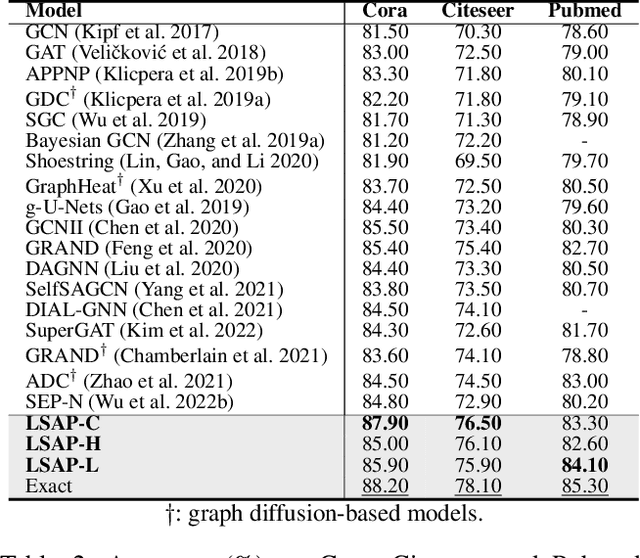
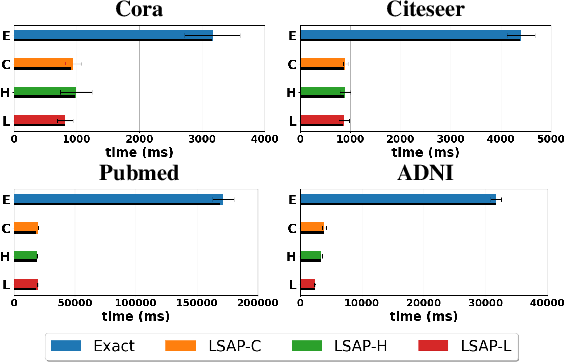
Abstract:Various Graph Neural Networks (GNNs) have been successful in analyzing data in non-Euclidean spaces, however, they have limitations such as oversmoothing, i.e., information becomes excessively averaged as the number of hidden layers increases. The issue stems from the intrinsic formulation of conventional graph convolution where the nodal features are aggregated from a direct neighborhood per layer across the entire nodes in the graph. As setting different number of hidden layers per node is infeasible, recent works leverage a diffusion kernel to redefine the graph structure and incorporate information from farther nodes. Unfortunately, such approaches suffer from heavy diagonalization of a graph Laplacian or learning a large transform matrix. In this regards, we propose a diffusion learning framework, where the range of feature aggregation is controlled by the scale of a diffusion kernel. For efficient computation, we derive closed-form derivatives of approximations of the graph convolution with respect to the scale, so that node-wise range can be adaptively learned. With a downstream classifier, the entire framework is made trainable in an end-to-end manner. Our model is tested on various standard datasets for node-wise classification for the state-of-the-art performance, and it is also validated on a real-world brain network data for graph classifications to demonstrate its practicality for Alzheimer classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge