Mustafa Dere
Modality-Agnostic Style Transfer for Holistic Feature Imputation
Mar 03, 2025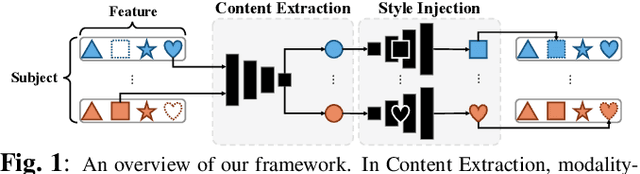
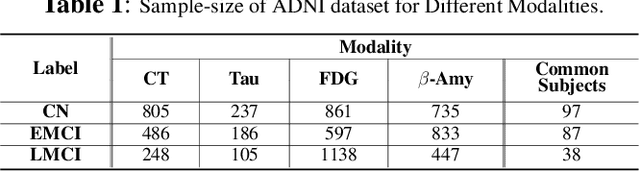
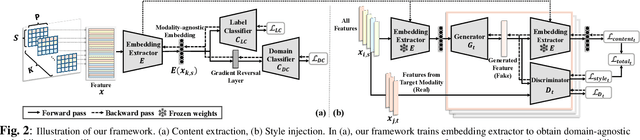
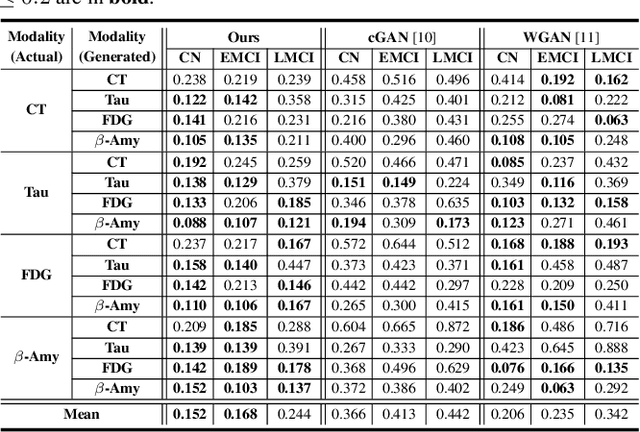
Abstract:Characterizing a preclinical stage of Alzheimer's Disease (AD) via single imaging is difficult as its early symptoms are quite subtle. Therefore, many neuroimaging studies are curated with various imaging modalities, e.g., MRI and PET, however, it is often challenging to acquire all of them from all subjects and missing data become inevitable. In this regards, in this paper, we propose a framework that generates unobserved imaging measures for specific subjects using their existing measures, thereby reducing the need for additional examinations. Our framework transfers modality-specific style while preserving AD-specific content. This is done by domain adversarial training that preserves modality-agnostic but AD-specific information, while a generative adversarial network adds an indistinguishable modality-specific style. Our proposed framework is evaluated on the Alzheimer's Disease Neuroimaging Initiative (ADNI) study and compared with other imputation methods in terms of generated data quality. Small average Cohen's $d$ $< 0.19$ between our generated measures and real ones suggests that the synthetic data are practically usable regardless of their modality type.
Learning Covariance-Based Multi-Scale Representation of Neuroimaging Measures for Alzheimer Classification
Mar 03, 2025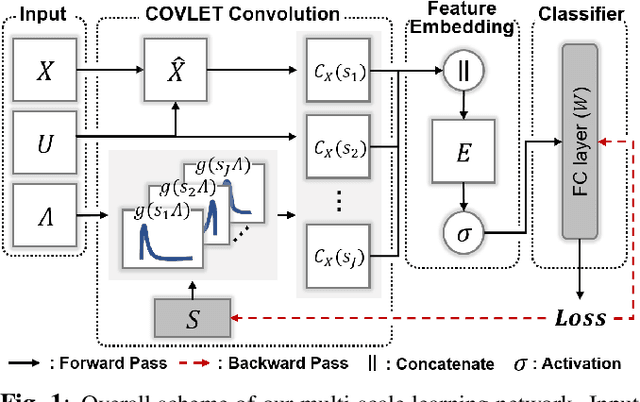
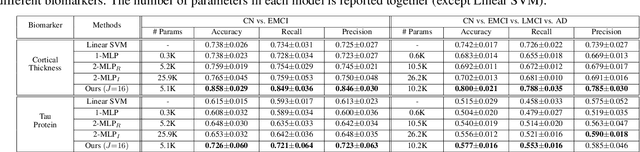
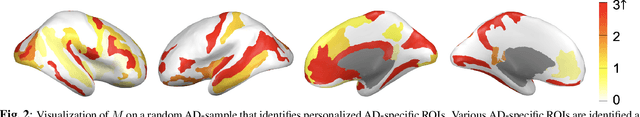
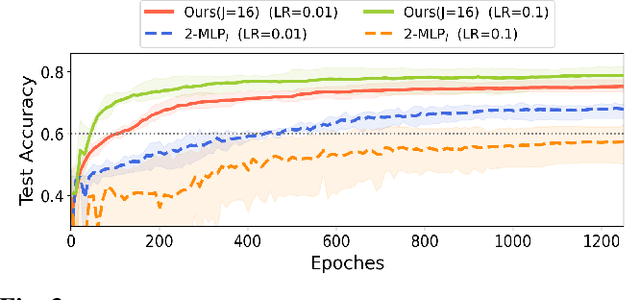
Abstract:Stacking excessive layers in DNN results in highly underdetermined system when training samples are limited, which is very common in medical applications. In this regard, we present a framework capable of deriving an efficient high-dimensional space with reasonable increase in model size. This is done by utilizing a transform (i.e., convolution) that leverages scale-space theory with covariance structure. The overall model trains on this transform together with a downstream classifier (i.e., Fully Connected layer) to capture the optimal multi-scale representation of the original data which corresponds to task-specific components in a dual space. Experiments on neuroimaging measures from Alzheimer's Disease Neuroimaging Initiative (ADNI) study show that our model performs better and converges faster than conventional models even when the model size is significantly reduced. The trained model is made interpretable using gradient information over the multi-scale transform to delineate personalized AD-specific regions in the brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge