Jaeyoon Sim
OCL: Ordinal Contrastive Learning for Imputating Features with Progressive Labels
Mar 03, 2025Abstract:Accurately discriminating progressive stages of Alzheimer's Disease (AD) is crucial for early diagnosis and prevention. It often involves multiple imaging modalities to understand the complex pathology of AD, however, acquiring a complete set of images is challenging due to high cost and burden for subjects. In the end, missing data become inevitable which lead to limited sample-size and decrease in precision in downstream analyses. To tackle this challenge, we introduce a holistic imaging feature imputation method that enables to leverage diverse imaging features while retaining all subjects. The proposed method comprises two networks: 1) An encoder to extract modality-independent embeddings and 2) A decoder to reconstruct the original measures conditioned on their imaging modalities. The encoder includes a novel {\em ordinal contrastive loss}, which aligns samples in the embedding space according to the progression of AD. We also maximize modality-wise coherence of embeddings within each subject, in conjunction with domain adversarial training algorithms, to further enhance alignment between different imaging modalities. The proposed method promotes our holistic imaging feature imputation across various modalities in the shared embedding space. In the experiments, we show that our networks deliver favorable results for statistical analysis and classification against imputation baselines with Alzheimer's Disease Neuroimaging Initiative (ADNI) study.
Modality-Agnostic Style Transfer for Holistic Feature Imputation
Mar 03, 2025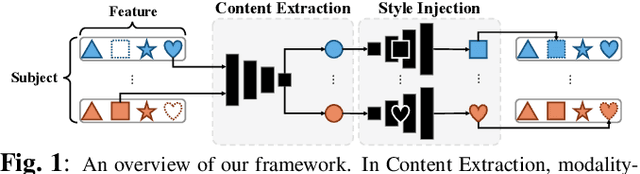
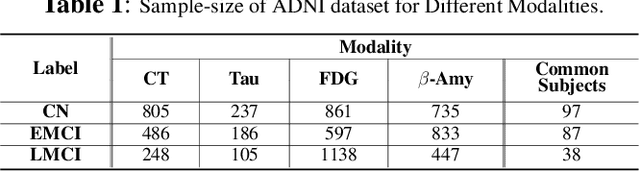
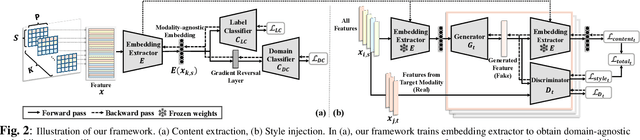
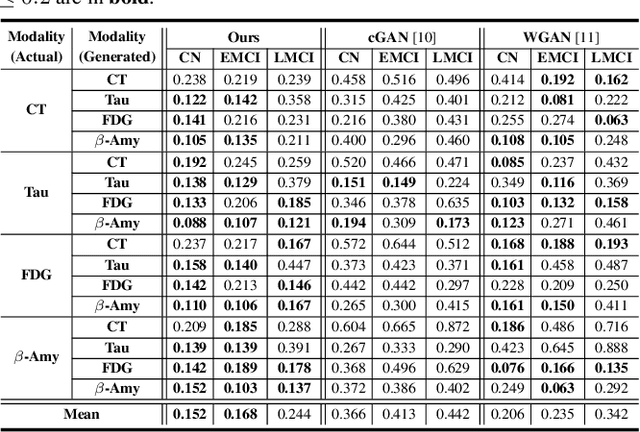
Abstract:Characterizing a preclinical stage of Alzheimer's Disease (AD) via single imaging is difficult as its early symptoms are quite subtle. Therefore, many neuroimaging studies are curated with various imaging modalities, e.g., MRI and PET, however, it is often challenging to acquire all of them from all subjects and missing data become inevitable. In this regards, in this paper, we propose a framework that generates unobserved imaging measures for specific subjects using their existing measures, thereby reducing the need for additional examinations. Our framework transfers modality-specific style while preserving AD-specific content. This is done by domain adversarial training that preserves modality-agnostic but AD-specific information, while a generative adversarial network adds an indistinguishable modality-specific style. Our proposed framework is evaluated on the Alzheimer's Disease Neuroimaging Initiative (ADNI) study and compared with other imputation methods in terms of generated data quality. Small average Cohen's $d$ $< 0.19$ between our generated measures and real ones suggests that the synthetic data are practically usable regardless of their modality type.
Learning to Approximate Adaptive Kernel Convolution on Graphs
Jan 22, 2024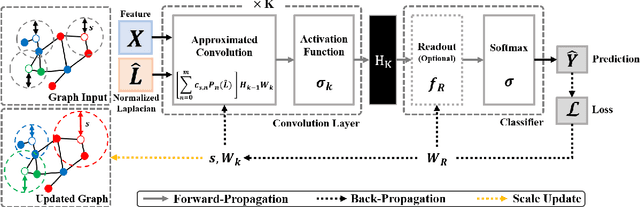
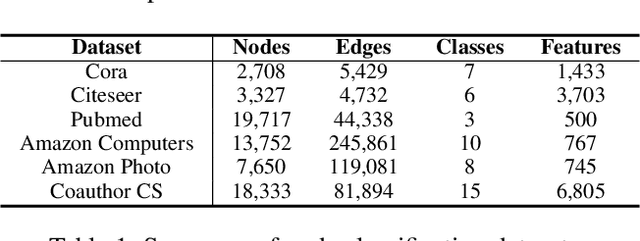
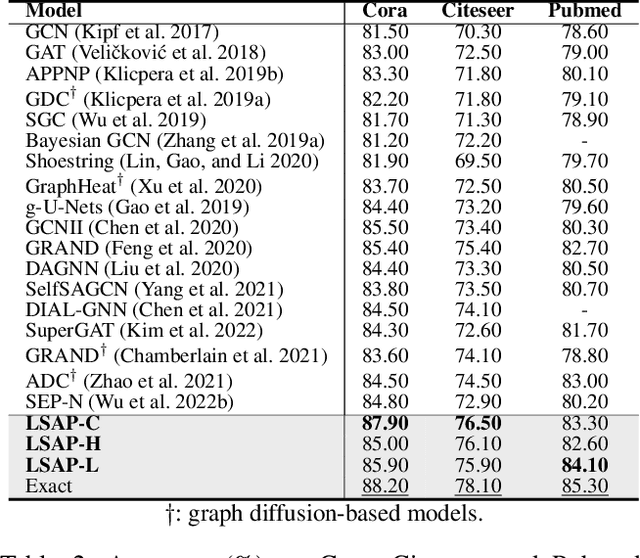
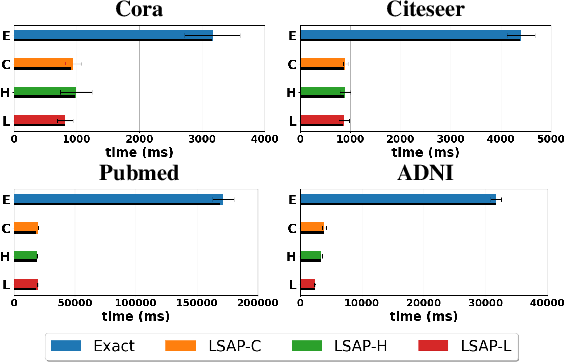
Abstract:Various Graph Neural Networks (GNNs) have been successful in analyzing data in non-Euclidean spaces, however, they have limitations such as oversmoothing, i.e., information becomes excessively averaged as the number of hidden layers increases. The issue stems from the intrinsic formulation of conventional graph convolution where the nodal features are aggregated from a direct neighborhood per layer across the entire nodes in the graph. As setting different number of hidden layers per node is infeasible, recent works leverage a diffusion kernel to redefine the graph structure and incorporate information from farther nodes. Unfortunately, such approaches suffer from heavy diagonalization of a graph Laplacian or learning a large transform matrix. In this regards, we propose a diffusion learning framework, where the range of feature aggregation is controlled by the scale of a diffusion kernel. For efficient computation, we derive closed-form derivatives of approximations of the graph convolution with respect to the scale, so that node-wise range can be adaptively learned. With a downstream classifier, the entire framework is made trainable in an end-to-end manner. Our model is tested on various standard datasets for node-wise classification for the state-of-the-art performance, and it is also validated on a real-world brain network data for graph classifications to demonstrate its practicality for Alzheimer classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge