Gernot Plank
Explainable Deep Learning-based Classification of Wolff-Parkinson-White Electrocardiographic Signals
Nov 08, 2025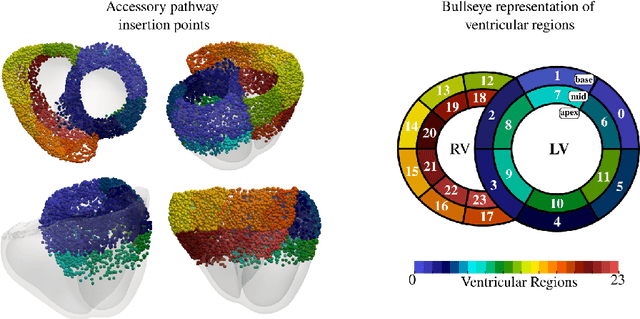
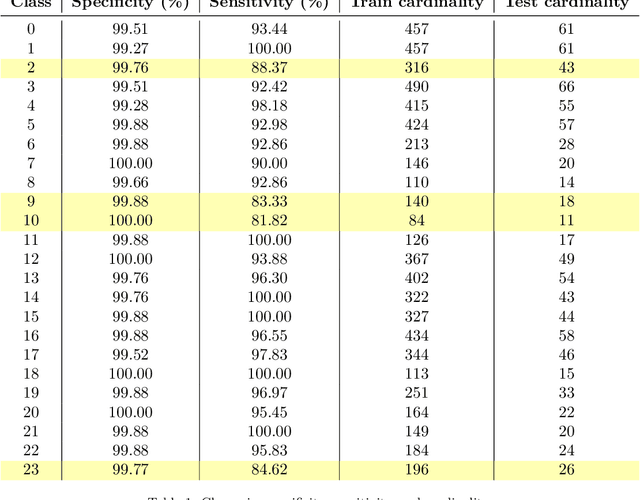
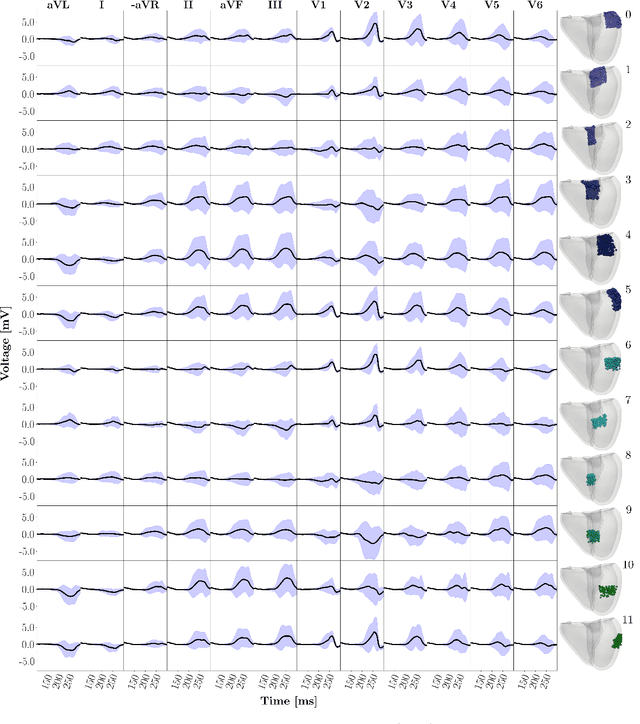
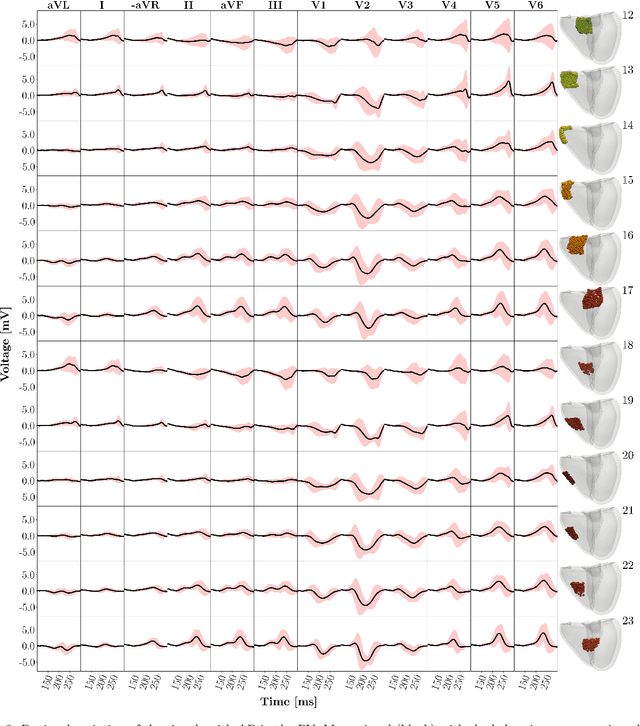
Abstract:Wolff-Parkinson-White (WPW) syndrome is a cardiac electrophysiology (EP) disorder caused by the presence of an accessory pathway (AP) that bypasses the atrioventricular node, faster ventricular activation rate, and provides a substrate for atrio-ventricular reentrant tachycardia (AVRT). Accurate localization of the AP is critical for planning and guiding catheter ablation procedures. While traditional diagnostic tree (DT) methods and more recent machine learning (ML) approaches have been proposed to predict AP location from surface electrocardiogram (ECG), they are often constrained by limited anatomical localization resolution, poor interpretability, and the use of small clinical datasets. In this study, we present a Deep Learning (DL) model for the localization of single manifest APs across 24 cardiac regions, trained on a large, physiologically realistic database of synthetic ECGs generated using a personalized virtual heart model. We also integrate eXplainable Artificial Intelligence (XAI) methods, Guided Backpropagation, Grad-CAM, and Guided Grad-CAM, into the pipeline. This enables interpretation of DL decision-making and addresses one of the main barriers to clinical adoption: lack of transparency in ML predictions. Our model achieves localization accuracy above 95%, with a sensitivity of 94.32% and specificity of 99.78%. XAI outputs are physiologically validated against known depolarization patterns, and a novel index is introduced to identify the most informative ECG leads for AP localization. Results highlight lead V2 as the most critical, followed by aVF, V1, and aVL. This work demonstrates the potential of combining cardiac digital twins with explainable DL to enable accurate, transparent, and non-invasive AP localization.
Augmentation-based Domain Generalization and Joint Training from Multiple Source Domains for Whole Heart Segmentation
Aug 06, 2025Abstract:As the leading cause of death worldwide, cardiovascular diseases motivate the development of more sophisticated methods to analyze the heart and its substructures from medical images like Computed Tomography (CT) and Magnetic Resonance (MR). Semantic segmentations of important cardiac structures that represent the whole heart are useful to assess patient-specific cardiac morphology and pathology. Furthermore, accurate semantic segmentations can be used to generate cardiac digital twin models which allows e.g. electrophysiological simulation and personalized therapy planning. Even though deep learning-based methods for medical image segmentation achieved great advancements over the last decade, retaining good performance under domain shift -- i.e. when training and test data are sampled from different data distributions -- remains challenging. In order to perform well on domains known at training-time, we employ a (1) balanced joint training approach that utilizes CT and MR data in equal amounts from different source domains. Further, aiming to alleviate domain shift towards domains only encountered at test-time, we rely on (2) strong intensity and spatial augmentation techniques to greatly diversify the available training data. Our proposed whole heart segmentation method, a 5-fold ensemble with our contributions, achieves the best performance for MR data overall and a performance similar to the best performance for CT data when compared to a model trained solely on CT. With 93.33% DSC and 0.8388 mm ASSD for CT and 89.30% DSC and 1.2411 mm ASSD for MR data, our method demonstrates great potential to efficiently obtain accurate semantic segmentations from which patient-specific cardiac twin models can be generated.
LA-CaRe-CNN: Cascading Refinement CNN for Left Atrial Scar Segmentation
Aug 06, 2025Abstract:Atrial fibrillation (AF) represents the most prevalent type of cardiac arrhythmia for which treatment may require patients to undergo ablation therapy. In this surgery cardiac tissues are locally scarred on purpose to prevent electrical signals from causing arrhythmia. Patient-specific cardiac digital twin models show great potential for personalized ablation therapy, however, they demand accurate semantic segmentation of healthy and scarred tissue typically obtained from late gadolinium enhanced (LGE) magnetic resonance (MR) scans. In this work we propose the Left Atrial Cascading Refinement CNN (LA-CaRe-CNN), which aims to accurately segment the left atrium as well as left atrial scar tissue from LGE MR scans. LA-CaRe-CNN is a 2-stage CNN cascade that is trained end-to-end in 3D, where Stage 1 generates a prediction for the left atrium, which is then refined in Stage 2 in conjunction with the original image information to obtain a prediction for the left atrial scar tissue. To account for domain shift towards domains unknown during training, we employ strong intensity and spatial augmentation to increase the diversity of the training dataset. Our proposed method based on a 5-fold ensemble achieves great segmentation results, namely, 89.21% DSC and 1.6969 mm ASSD for the left atrium, as well as 64.59% DSC and 91.80% G-DSC for the more challenging left atrial scar tissue. Thus, segmentations obtained through LA-CaRe-CNN show great potential for the generation of patient-specific cardiac digital twin models and downstream tasks like personalized targeted ablation therapy to treat AF.
Physics-informed Neural Network Estimation of Material Properties in Soft Tissue Nonlinear Biomechanical Models
Dec 20, 2023Abstract:The development of biophysical models for clinical applications is rapidly advancing in the research community, thanks to their predictive nature and their ability to assist the interpretation of clinical data. However, high-resolution and accurate multi-physics computational models are computationally expensive and their personalisation involves fine calibration of a large number of parameters, which may be space-dependent, challenging their clinical translation. In this work, we propose a new approach which relies on the combination of physics-informed neural networks (PINNs) with three-dimensional soft tissue nonlinear biomechanical models, capable of reconstructing displacement fields and estimating heterogeneous patient-specific biophysical properties. The proposed learning algorithm encodes information from a limited amount of displacement and, in some cases, strain data, that can be routinely acquired in the clinical setting, and combines it with the physics of the problem, represented by a mathematical model based on partial differential equations, to regularise the problem and improve its convergence properties. Several benchmarks are presented to show the accuracy and robustness of the proposed method and its great potential to enable the robust and effective identification of patient-specific, heterogeneous physical properties, s.a. tissue stiffness properties. In particular, we demonstrate the capability of the PINN to detect the presence, location and severity of scar tissue, which is beneficial to develop personalised simulation models for disease diagnosis, especially for cardiac applications.
CaRe-CNN: Cascading Refinement CNN for Myocardial Infarct Segmentation with Microvascular Obstructions
Dec 19, 2023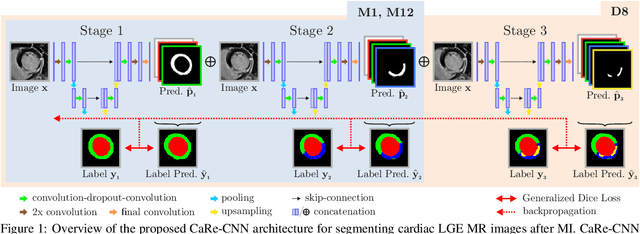
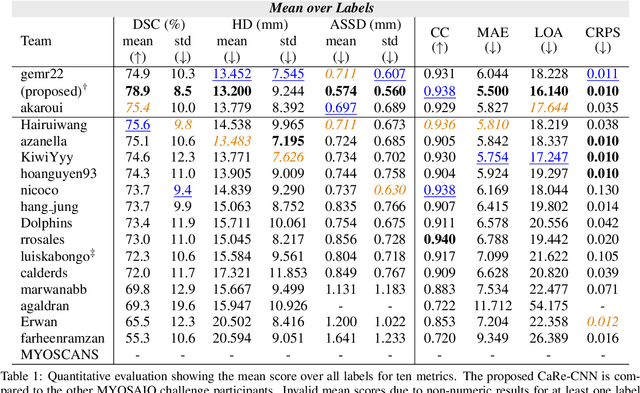
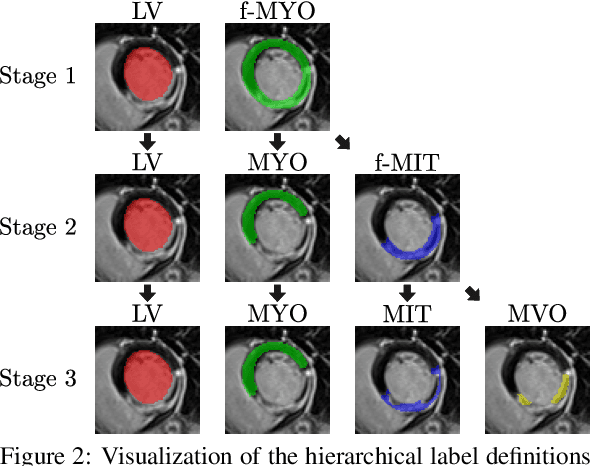
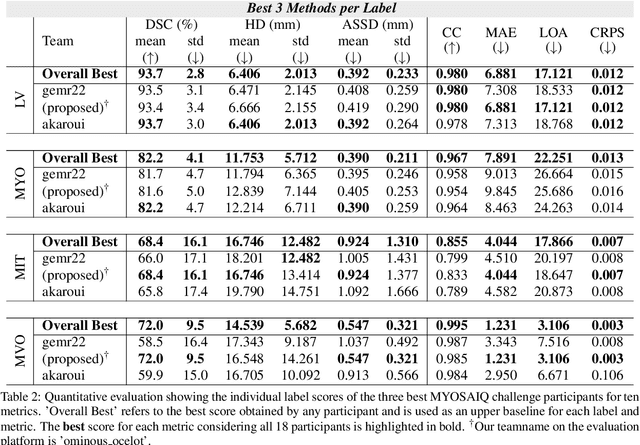
Abstract:Late gadolinium enhanced (LGE) magnetic resonance (MR) imaging is widely established to assess the viability of myocardial tissue of patients after acute myocardial infarction (MI). We propose the Cascading Refinement CNN (CaRe-CNN), which is a fully 3D, end-to-end trained, 3-stage CNN cascade that exploits the hierarchical structure of such labeled cardiac data. Throughout the three stages of the cascade, the label definition changes and CaRe-CNN learns to gradually refine its intermediate predictions accordingly. Furthermore, to obtain more consistent qualitative predictions, we propose a series of post-processing steps that take anatomical constraints into account. Our CaRe-CNN was submitted to the FIMH 2023 MYOSAIQ challenge, where it ranked second out of 18 participating teams. CaRe-CNN showed great improvements most notably when segmenting the difficult but clinically most relevant myocardial infarct tissue (MIT) as well as microvascular obstructions (MVO). When computing the average scores over all labels, our method obtained the best score in eight out of ten metrics. Thus, accurate cardiac segmentation after acute MI via our CaRe-CNN allows generating patient-specific models of the heart serving as an important step towards personalized medicine.
MedalCare-XL: 16,900 healthy and pathological 12 lead ECGs obtained through electrophysiological simulations
Nov 29, 2022Abstract:Mechanistic cardiac electrophysiology models allow for personalized simulations of the electrical activity in the heart and the ensuing electrocardiogram (ECG) on the body surface. As such, synthetic signals possess known ground truth labels of the underlying disease and can be employed for validation of machine learning ECG analysis tools in addition to clinical signals. Recently, synthetic ECGs were used to enrich sparse clinical data or even replace them completely during training leading to improved performance on real-world clinical test data. We thus generated a novel synthetic database comprising a total of 16,900 12 lead ECGs based on electrophysiological simulations equally distributed into healthy control and 7 pathology classes. The pathological case of myocardial infraction had 6 sub-classes. A comparison of extracted features between the virtual cohort and a publicly available clinical ECG database demonstrated that the synthetic signals represent clinical ECGs for healthy and pathological subpopulations with high fidelity. The ECG database is split into training, validation, and test folds for development and objective assessment of novel machine learning algorithms.
Comparison of propagation models and forward calculation methods on cellular, tissue and organ scale atrial electrophysiology
Mar 15, 2022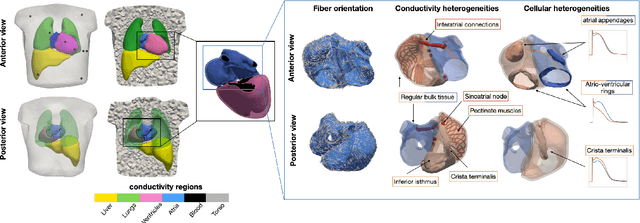
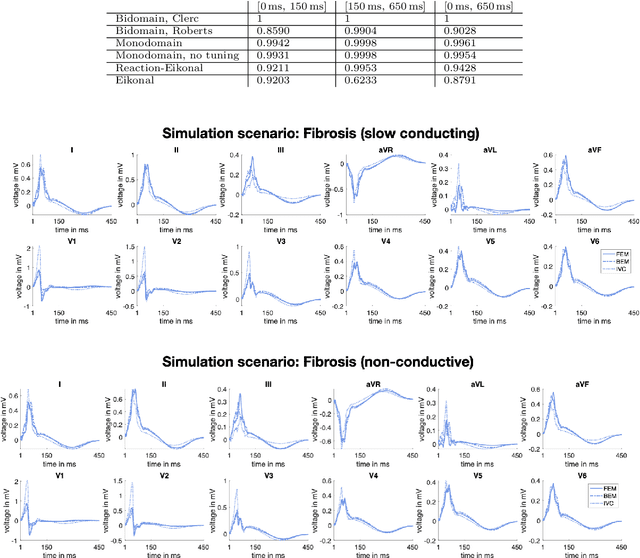
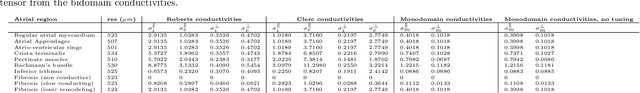
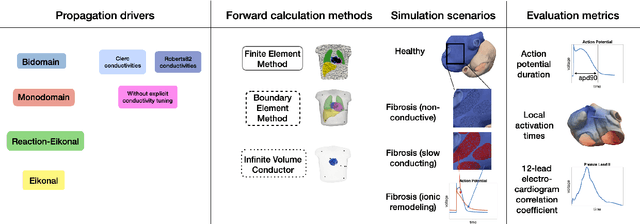
Abstract:Objective: The bidomain model and the finite element method are an established standard to mathematically describe cardiac electrophysiology, but are both suboptimal choices for fast and large-scale simulations due to high computational costs. We investigate to what extent simplified approaches for propagation models (monodomain, reaction-eikonal and eikonal) and forward calculation (boundary element and infinite volume conductor) deliver markedly accelerated, yet physiologically accurate simulation results in atrial electrophysiology. Methods: We compared action potential durations, local activation times (LATs), and electrocardiograms (ECGs) for sinus rhythm simulations on healthy and fibrotically infiltrated atrial models. Results: All simplified model solutions yielded LATs and P waves in accurate accordance with the bidomain results. Only for the eikonal model with pre-computed action potential templates shifted in time to derive transmembrane voltages, repolarization behavior notably deviated from the bidomain results. ECGs calculated with the boundary element method were characterized by correlation coefficients >0.9 compared to the finite element method. The infinite volume conductor method led to lower correlation coefficients caused predominantly by systematic overestimations of P wave amplitudes in the precordial leads. Conclusion: Our results demonstrate that the eikonal model yields accurate LATs and combined with the boundary element method precise ECGs compared to markedly more expensive full bidomain simulations. However, for an accurate representation of atrial repolarization dynamics, diffusion terms must be accounted for in simplified models. Significance: Simulations of atrial LATs and ECGs can be notably accelerated to clinically feasible time frames at high accuracy by resorting to the eikonal and boundary element methods.
Physics-informed neural networks to learn cardiac fiber orientation from multiple electroanatomical maps
Feb 01, 2022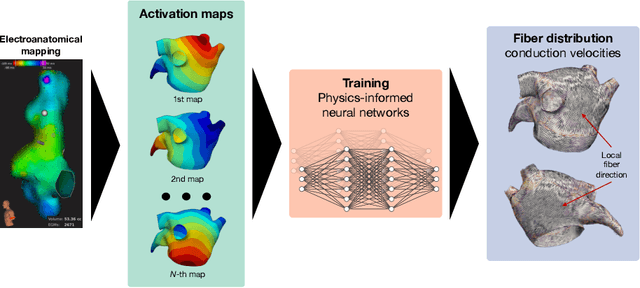

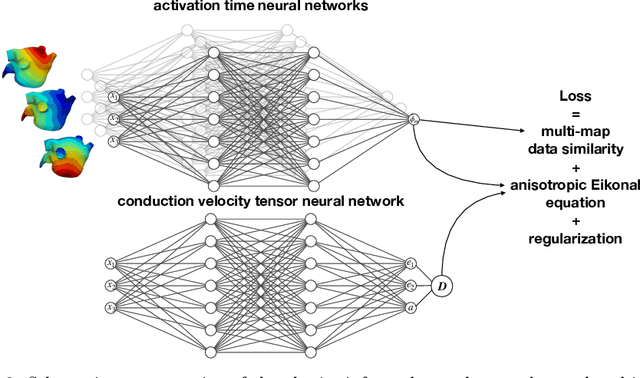
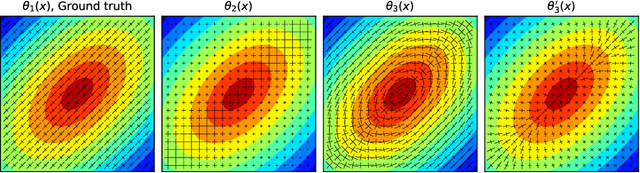
Abstract:We propose FiberNet, a method to estimate in-vivo the cardiac fiber architecture of the human atria from multiple catheter recordings of the electrical activation. Cardiac fibers play a central rolein the electro-mechanical function of the heart, yet they aredifficult to determine in-vivo, and hence rarely truly patient-specificin existing cardiac models.FiberNet learns the fibers arrangement by solvingan inverse problem with physics-informed neural networks. The inverse problem amounts to identifyingthe conduction velocity tensor of a cardiac propagation modelfrom a set of sparse activation maps. The use of multiple mapsenables the simultaneous identification of all the componentsof the conduction velocity tensor, including the local fiber angle.We extensively test FiberNet on synthetic 2-D and 3-D examples, diffusion tensor fibers, and a patient-specific case. We show that 3 maps are sufficient to accurately capture the fibers, also in thepresence of noise. With fewer maps, the role of regularization becomesprominent. Moreover, we show that the fitted model can robustlyreproduce unseen activation maps. We envision that FiberNet will help the creation of patient-specific models for personalized medicine.The full code is available at http://github.com/fsahli/FiberNet.
Smoothness and continuity of cost functionals for ECG mismatch computation
Jan 12, 2022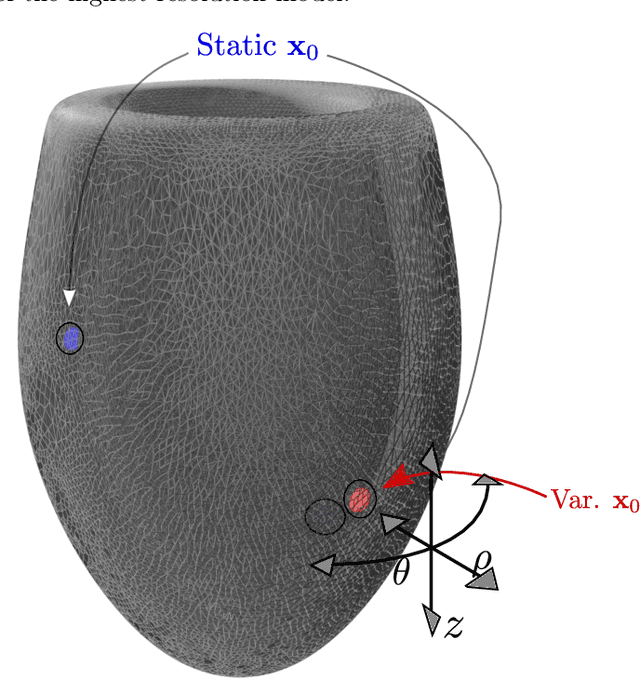
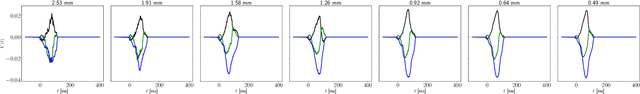
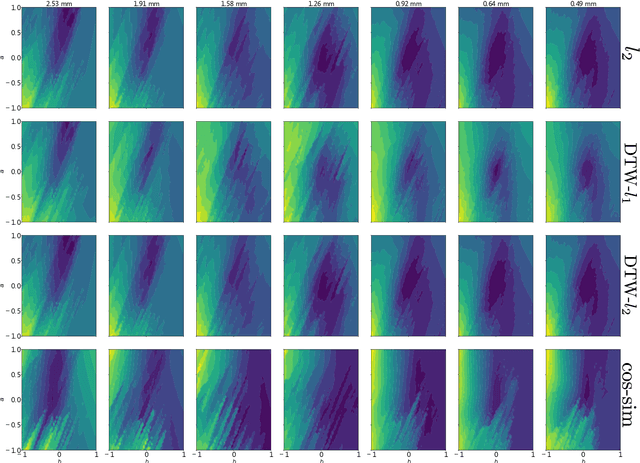
Abstract:The field of cardiac electrophysiology tries to abstract, describe and finally model the electrical characteristics of a heartbeat. With recent advances in cardiac electrophysiology, models have become more powerful and descriptive as ever. However, to advance to the field of inverse electrophysiological modeling, i.e. creating models from electrical measurements such as the ECG, the less investigated field of smoothness of the simulated ECGs w.r.t. model parameters need to be further explored. The present paper discusses smoothness in terms of the whole pipeline which describes how from physiological parameters, we arrive at the simulated ECG. Employing such a pipeline, we create a test-bench of a simplified idealized left ventricle model and demonstrate the most important factors for efficient inverse modeling through smooth cost functionals. Such knowledge will be important for designing and creating inverse models in future optimization and machine learning methods.
Learning atrial fiber orientations and conductivity tensors from intracardiac maps using physics-informed neural networks
Feb 22, 2021
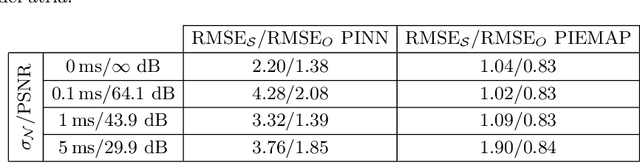

Abstract:Electroanatomical maps are a key tool in the diagnosis and treatment of atrial fibrillation. Current approaches focus on the activation times recorded. However, more information can be extracted from the available data. The fibers in cardiac tissue conduct the electrical wave faster, and their direction could be inferred from activation times. In this work, we employ a recently developed approach, called physics informed neural networks, to learn the fiber orientations from electroanatomical maps, taking into account the physics of the electrical wave propagation. In particular, we train the neural network to weakly satisfy the anisotropic eikonal equation and to predict the measured activation times. We use a local basis for the anisotropic conductivity tensor, which encodes the fiber orientation. The methodology is tested both in a synthetic example and for patient data. Our approach shows good agreement in both cases and it outperforms a state of the art method in the patient data. The results show a first step towards learning the fiber orientations from electroanatomical maps with physics-informed neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge