Tobias Schäffter
Physikalisch Technische Bundesanstalt, Abbestrasse 2-12, 10587 Berlin, Germany, Technical University of Berlin, Department of Medical Engineering, Dovestrasse 6, Berlin, Germany
The METRIC-framework for assessing data quality for trustworthy AI in medicine: a systematic review
Feb 21, 2024Abstract:The adoption of machine learning (ML) and, more specifically, deep learning (DL) applications into all major areas of our lives is underway. The development of trustworthy AI is especially important in medicine due to the large implications for patients' lives. While trustworthiness concerns various aspects including ethical, technical and privacy requirements, we focus on the importance of data quality (training/test) in DL. Since data quality dictates the behaviour of ML products, evaluating data quality will play a key part in the regulatory approval of medical AI products. We perform a systematic review following PRISMA guidelines using the databases PubMed and ACM Digital Library. We identify 2362 studies, out of which 62 records fulfil our eligibility criteria. From this literature, we synthesise the existing knowledge on data quality frameworks and combine it with the perspective of ML applications in medicine. As a result, we propose the METRIC-framework, a specialised data quality framework for medical training data comprising 15 awareness dimensions, along which developers of medical ML applications should investigate a dataset. This knowledge helps to reduce biases as a major source of unfairness, increase robustness, facilitate interpretability and thus lays the foundation for trustworthy AI in medicine. Incorporating such systematic assessment of medical datasets into regulatory approval processes has the potential to accelerate the approval of ML products and builds the basis for new standards.
Simulation of acquisition shifts in T2 Flair MR images to stress test AI segmentation networks
Nov 03, 2023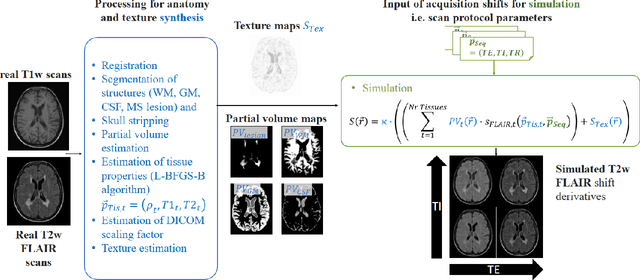

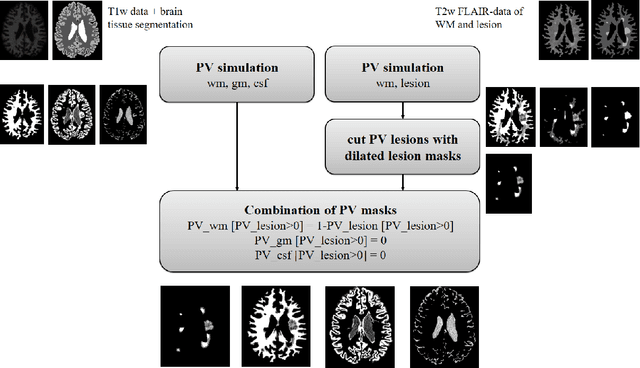
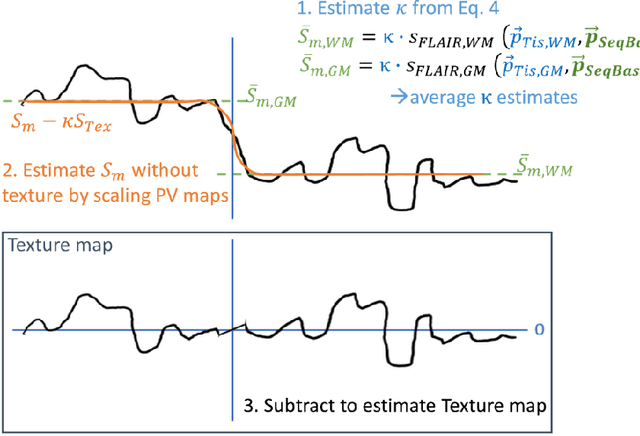
Abstract:Purpose: To provide a simulation framework for routine neuroimaging test data, which allows for "stress testing" of deep segmentation networks against acquisition shifts that commonly occur in clinical practice for T2 weighted (T2w) fluid attenuated inversion recovery (FLAIR) Magnetic Resonance Imaging (MRI) protocols. Approach: The approach simulates "acquisition shift derivatives" of MR images based on MR signal equations. Experiments comprise the validation of the simulated images by real MR scans and example stress tests on state-of-the-art MS lesion segmentation networks to explore a generic model function to describe the F1 score in dependence of the contrast-affecting sequence parameters echo time (TE) and inversion time (TI). Results: The differences between real and simulated images range up to 19 % in gray and white matter for extreme parameter settings. For the segmentation networks under test the F1 score dependency on TE and TI can be well described by quadratic model functions (R^2 > 0.9). The coefficients of the model functions indicate that changes of TE have more influence on the model performance than TI. Conclusions: We show that these deviations are in the range of values as may be caused by erroneous or individual differences of relaxation times as described by literature. The coefficients of the F1 model function allow for quantitative comparison of the influences of TE and TI. Limitations arise mainly from tissues with the low baseline signal (like CSF) and when the protocol contains contrast-affecting measures that cannot be modelled due to missing information in the DICOM header.
MedalCare-XL: 16,900 healthy and pathological 12 lead ECGs obtained through electrophysiological simulations
Nov 29, 2022Abstract:Mechanistic cardiac electrophysiology models allow for personalized simulations of the electrical activity in the heart and the ensuing electrocardiogram (ECG) on the body surface. As such, synthetic signals possess known ground truth labels of the underlying disease and can be employed for validation of machine learning ECG analysis tools in addition to clinical signals. Recently, synthetic ECGs were used to enrich sparse clinical data or even replace them completely during training leading to improved performance on real-world clinical test data. We thus generated a novel synthetic database comprising a total of 16,900 12 lead ECGs based on electrophysiological simulations equally distributed into healthy control and 7 pathology classes. The pathological case of myocardial infraction had 6 sub-classes. A comparison of extracted features between the virtual cohort and a publicly available clinical ECG database demonstrated that the synthetic signals represent clinical ECGs for healthy and pathological subpopulations with high fidelity. The ECG database is split into training, validation, and test folds for development and objective assessment of novel machine learning algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge