Clare M. Tempany
Confidence Calibration and Predictive Uncertainty Estimation for Deep Medical Image Segmentation
Nov 29, 2019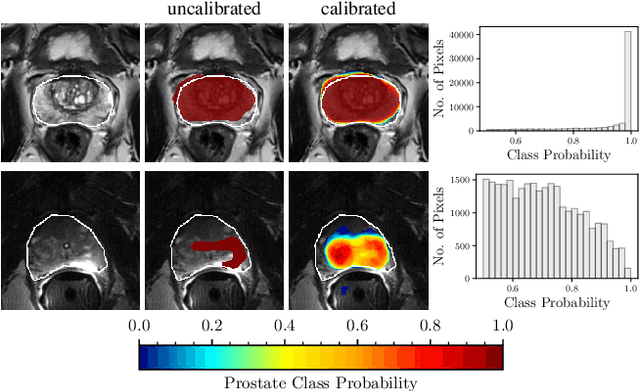
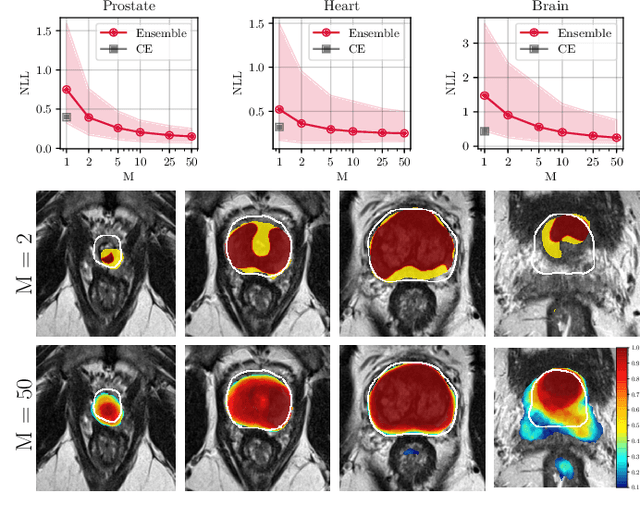
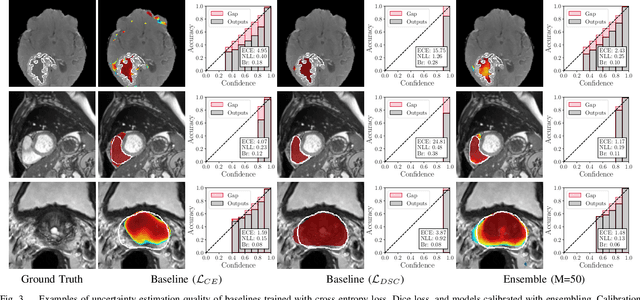
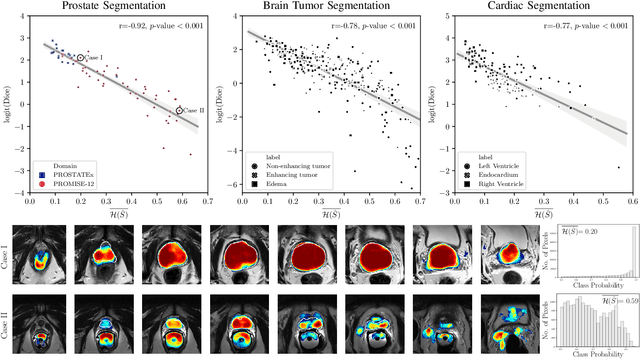
Abstract:Fully convolutional neural networks (FCNs), and in particular U-Nets, have achieved state-of-the-art results in semantic segmentation for numerous medical imaging applications. Moreover, batch normalization and Dice loss have been used successfully to stabilize and accelerate training. However, these networks are poorly calibrated i.e. they tend to produce overconfident predictions both in correct and erroneous classifications, making them unreliable and hard to interpret. In this paper, we study predictive uncertainty estimation in FCNs for medical image segmentation. We make the following contributions: 1) We systematically compare cross entropy loss with Dice loss in terms of segmentation quality and uncertainty estimation of FCNs; 2) We propose model ensembling for confidence calibration of the FCNs trained with batch normalization and Dice loss; 3) We assess the ability of calibrated FCNs to predict segmentation quality of structures and detect out-of-distribution test examples. We conduct extensive experiments across three medical image segmentation applications of the brain, the heart, and the prostate to evaluate our contributions. The results of this study offer considerable insight into the predictive uncertainty estimation and out-of-distribution detection in medical image segmentation and provide practical recipes for confidence calibration. Moreover, we consistently demonstrate that model ensembling improves confidence calibration.
Deep Information Theoretic Registration
Dec 31, 2018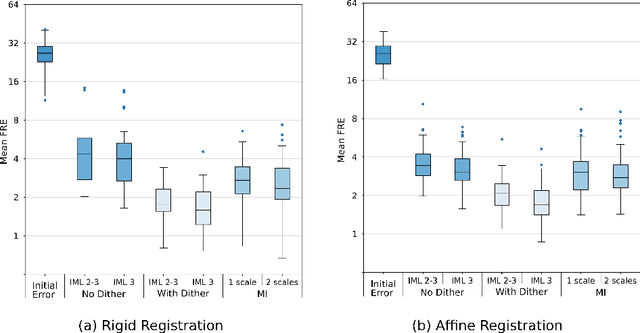
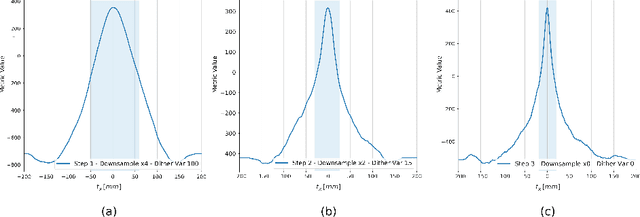
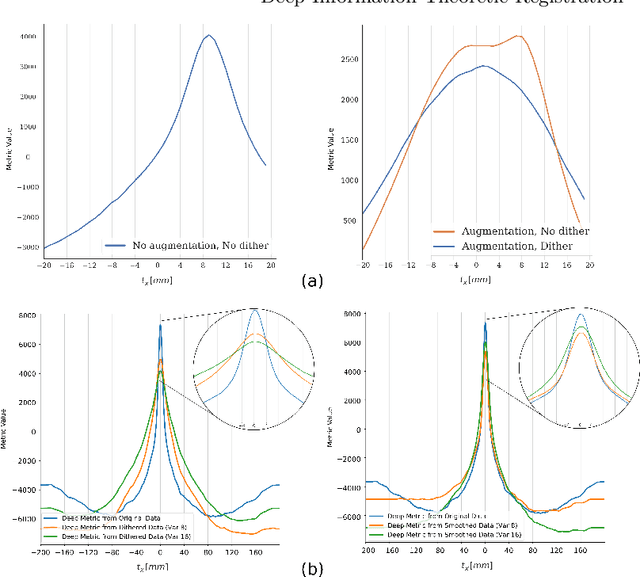
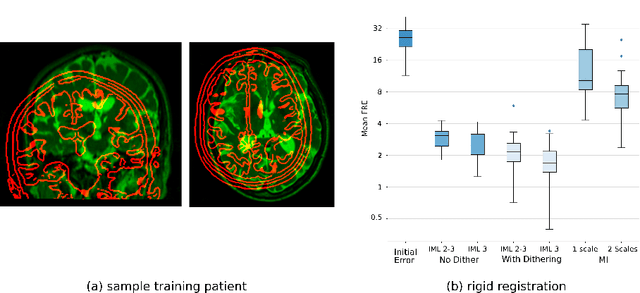
Abstract:This paper establishes an information theoretic framework for deep metric based image registration techniques. We show an exact equivalence between maximum profile likelihood and minimization of joint entropy, an important early information theoretic registration method. We further derive deep classifier-based metrics that can be used with iterated maximum likelihood to achieve Deep Information Theoretic Registration on patches rather than pixels. This alleviates a major shortcoming of previous information theoretic registration approaches, namely the implicit pixel-wise independence assumptions. Our proposed approach does not require well-registered training data; this brings previous fully supervised deep metric registration approaches to the realm of weak supervision. We evaluate our approach on several image registration tasks and show significantly better performance compared to mutual information, specifically when images have substantially different contrasts. This work enables general-purpose registration in applications where current methods are not successful.
Semi-Supervised Deep Metrics for Image Registration
Apr 04, 2018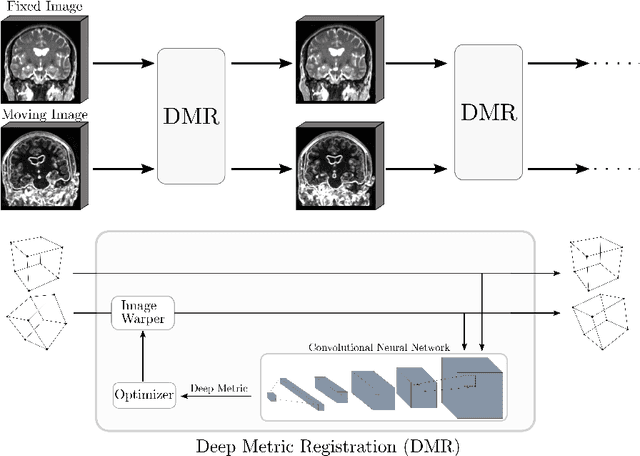
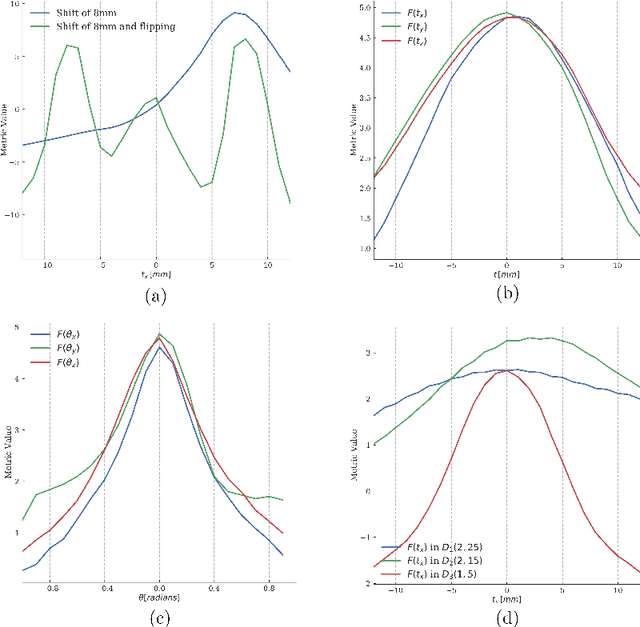
Abstract:Deep metrics have been shown effective as similarity measures in multi-modal image registration; however, the metrics are currently constructed from aligned image pairs in the training data. In this paper, we propose a strategy for learning such metrics from roughly aligned training data. Symmetrizing the data corrects bias in the metric that results from misalignment in the data (at the expense of increased variance), while random perturbations to the data, i.e. dithering, ensures that the metric has a single mode, and is amenable to registration by optimization. Evaluation is performed on the task of registration on separate unseen test image pairs. The results demonstrate the feasibility of learning a useful deep metric from substantially misaligned training data, in some cases the results are significantly better than from Mutual Information. Data augmentation via dithering is, therefore, an effective strategy for discharging the need for well-aligned training data; this brings deep metric registration from the realm of supervised to semi-supervised machine learning.
Transfer Learning for Domain Adaptation in MRI: Application in Brain Lesion Segmentation
Feb 25, 2017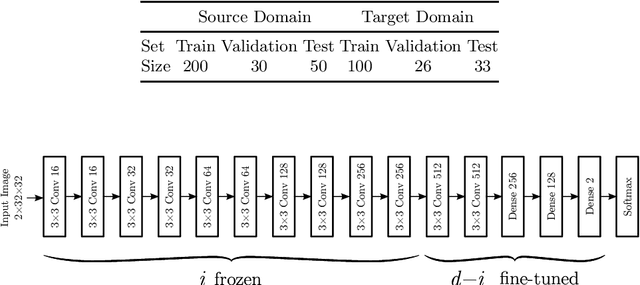
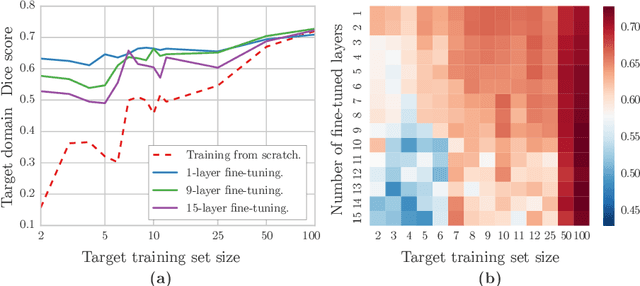
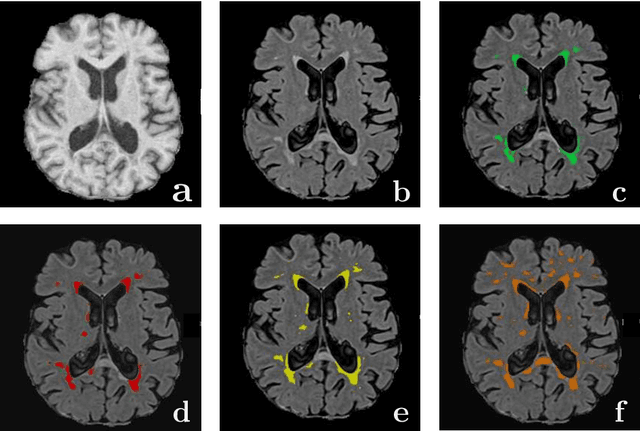
Abstract:Magnetic Resonance Imaging (MRI) is widely used in routine clinical diagnosis and treatment. However, variations in MRI acquisition protocols result in different appearances of normal and diseased tissue in the images. Convolutional neural networks (CNNs), which have shown to be successful in many medical image analysis tasks, are typically sensitive to the variations in imaging protocols. Therefore, in many cases, networks trained on data acquired with one MRI protocol, do not perform satisfactorily on data acquired with different protocols. This limits the use of models trained with large annotated legacy datasets on a new dataset with a different domain which is often a recurring situation in clinical settings. In this study, we aim to answer the following central questions regarding domain adaptation in medical image analysis: Given a fitted legacy model, 1) How much data from the new domain is required for a decent adaptation of the original network?; and, 2) What portion of the pre-trained model parameters should be retrained given a certain number of the new domain training samples? To address these questions, we conducted extensive experiments in white matter hyperintensity segmentation task. We trained a CNN on legacy MR images of brain and evaluated the performance of the domain-adapted network on the same task with images from a different domain. We then compared the performance of the model to the surrogate scenarios where either the same trained network is used or a new network is trained from scratch on the new dataset.The domain-adapted network tuned only by two training examples achieved a Dice score of 0.63 substantially outperforming a similar network trained on the same set of examples from scratch.
* 8 pages, 3 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge