Mehran Pesteie
Machine Learning Applications in Spine Biomechanics
Jan 10, 2024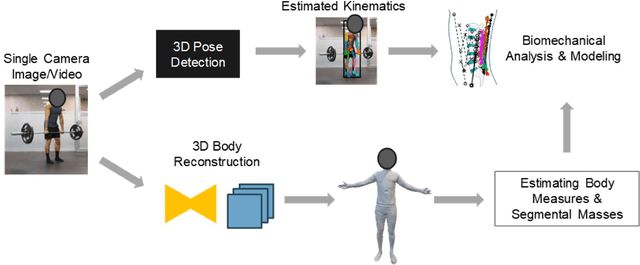

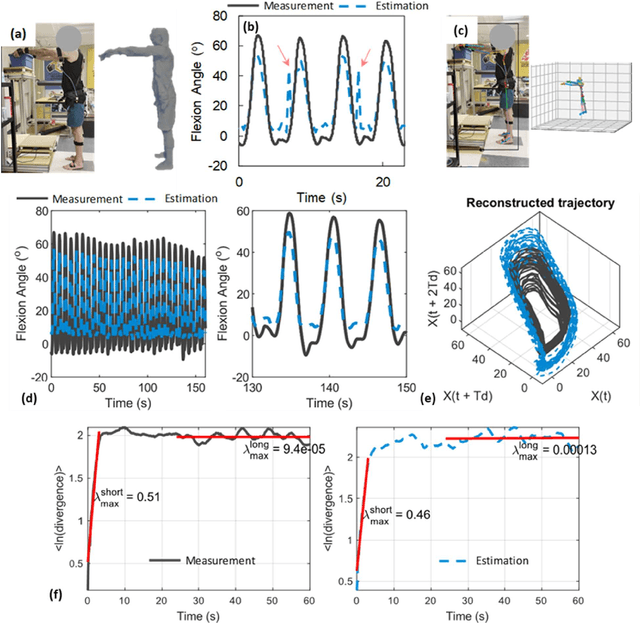
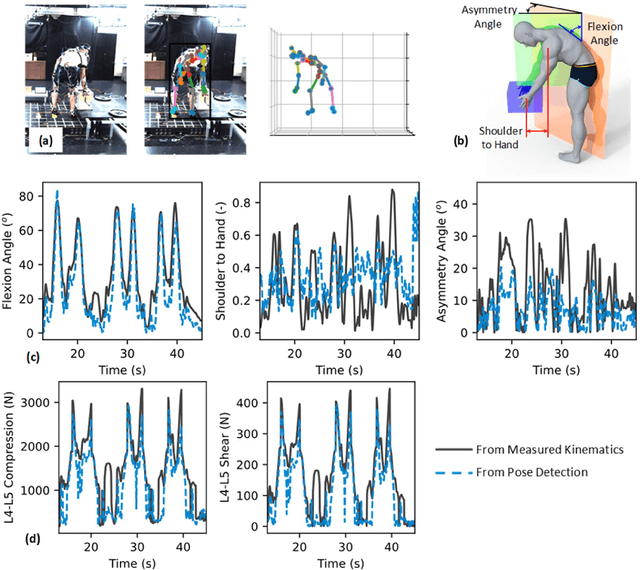
Abstract:Spine biomechanics is at a transformation with the advent and integration of machine learning and computer vision technologies. These novel techniques facilitate the estimation of 3D body shapes, anthropometrics, and kinematics from as simple as a single-camera image, making them more accessible and practical for a diverse range of applications. This study introduces a framework that merges these methodologies with traditional musculoskeletal modeling, enabling comprehensive analysis of spinal biomechanics during complex activities from a single camera. Additionally, we aim to evaluate their performance and limitations in spine biomechanics applications. The real-world applications explored in this study include assessment in workplace lifting, evaluation of whiplash injuries in car accidents, and biomechanical analysis in professional sports. Our results demonstrate potential and limitations of various algorithms in estimating body shape, kinematics, and conducting in-field biomechanical analyses. In industrial settings, the potential to utilize these new technologies for biomechanical risk assessments offers a pathway for preventive measures against back injuries. In sports activities, the proposed framework provides new opportunities for performance optimization, injury prevention, and rehabilitation. The application in forensic domain further underscores the wide-reaching implications of this technology. While certain limitations were identified, particularly in accuracy of predictions, complex interactions, and external load estimation, this study demonstrates their potential for advancement in spine biomechanics, heralding an optimistic future in both research and practical applications.
Variational Shape Completion for Virtual Planning of Jaw Reconstructive Surgery
Jul 15, 2019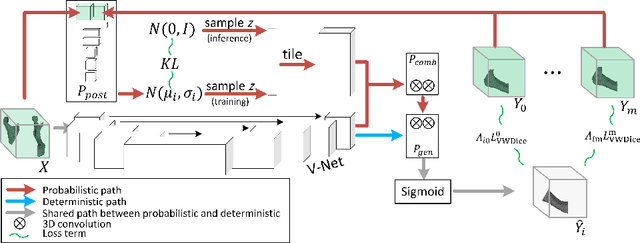
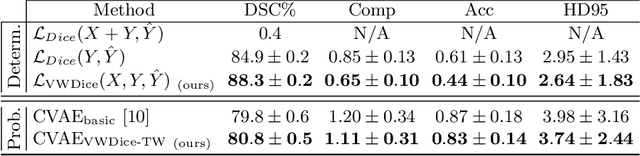
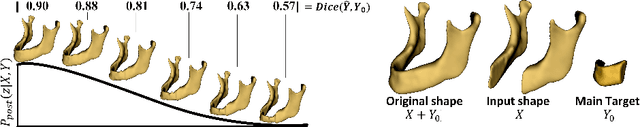
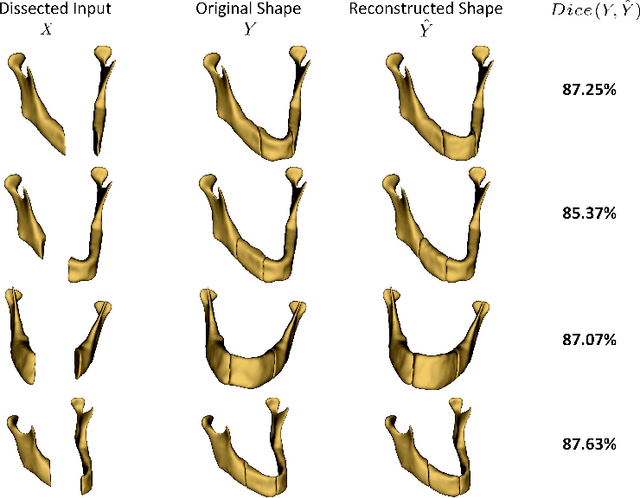
Abstract:The premorbid geometry of the mandible is of significant relevance in jaw reconstructive surgeries and occasionally unknown to the surgical team. In this paper, an optimization framework is introduced to train deep models for completion (reconstruction) of the missing segments of the bone based on the remaining healthy structure. To leverage the contextual information of the surroundings of the dissected region, the voxel-weighted Dice loss is introduced. To address the non-deterministic nature of the shape completion problem, we leverage a weighted multi-target probabilistic solution which is an extension to the conditional variational autoencoder (CVAE). This approach considers multiple targets as acceptable reconstructions, each weighted according to their conformity with the original shape. We quantify the performance gain of the proposed method against similar algorithms, including CVAE, where we report statistically significant improvements in both deterministic and probabilistic paradigms. The probabilistic model is also evaluated on its ability to generate anatomically relevant variations for the missing bone. As a unique aspect of this work, the model is tested on real surgical cases where the clinical relevancy of its reconstructions and their compliance with surgeon's virtual plan are demonstrated as necessary steps towards clinical adoption.
Deep Neural Maps
Oct 16, 2018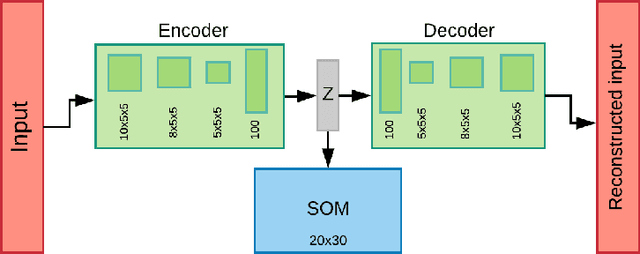
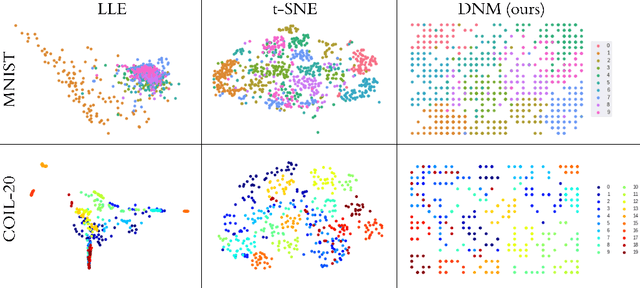
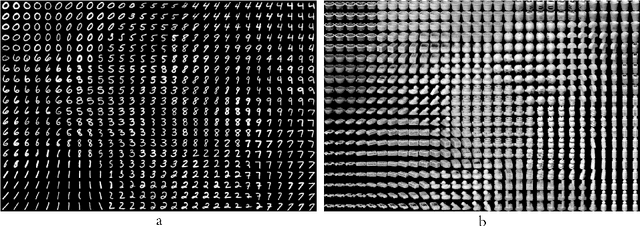
Abstract:We introduce a new unsupervised representation learning and visualization using deep convolutional networks and self organizing maps called Deep Neural Maps (DNM). DNM jointly learns an embedding of the input data and a mapping from the embedding space to a two-dimensional lattice. We compare visualizations of DNM with those of t-SNE and LLE on the MNIST and COIL-20 data sets. Our experiments show that the DNM can learn efficient representations of the input data, which reflects characteristics of each class. This is shown via back-projecting the neurons of the map on the data space.
Transfer Learning for Domain Adaptation in MRI: Application in Brain Lesion Segmentation
Feb 25, 2017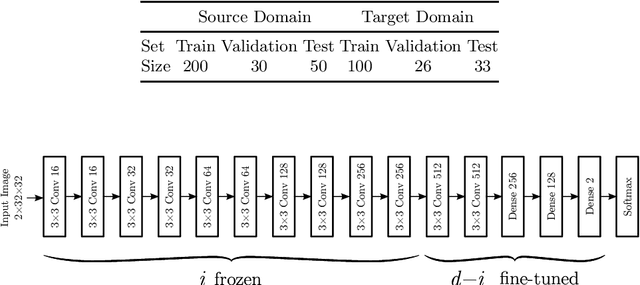
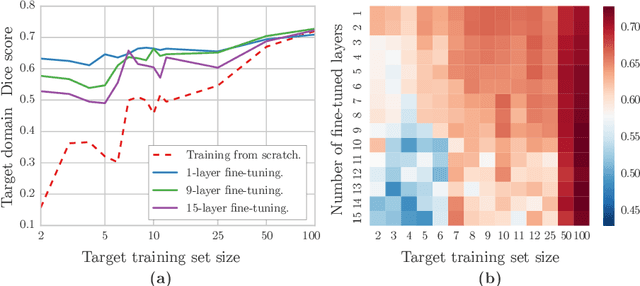
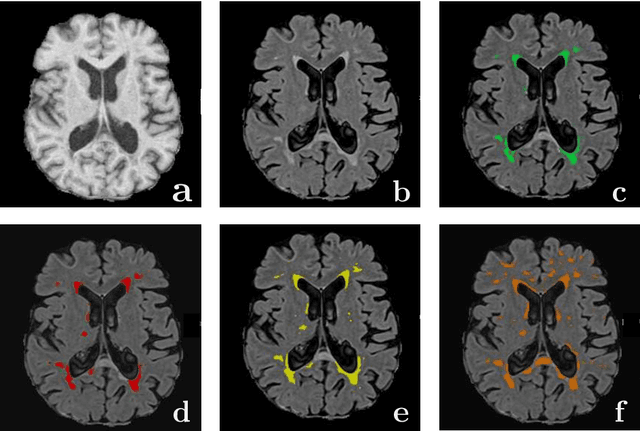
Abstract:Magnetic Resonance Imaging (MRI) is widely used in routine clinical diagnosis and treatment. However, variations in MRI acquisition protocols result in different appearances of normal and diseased tissue in the images. Convolutional neural networks (CNNs), which have shown to be successful in many medical image analysis tasks, are typically sensitive to the variations in imaging protocols. Therefore, in many cases, networks trained on data acquired with one MRI protocol, do not perform satisfactorily on data acquired with different protocols. This limits the use of models trained with large annotated legacy datasets on a new dataset with a different domain which is often a recurring situation in clinical settings. In this study, we aim to answer the following central questions regarding domain adaptation in medical image analysis: Given a fitted legacy model, 1) How much data from the new domain is required for a decent adaptation of the original network?; and, 2) What portion of the pre-trained model parameters should be retrained given a certain number of the new domain training samples? To address these questions, we conducted extensive experiments in white matter hyperintensity segmentation task. We trained a CNN on legacy MR images of brain and evaluated the performance of the domain-adapted network on the same task with images from a different domain. We then compared the performance of the model to the surrogate scenarios where either the same trained network is used or a new network is trained from scratch on the new dataset.The domain-adapted network tuned only by two training examples achieved a Dice score of 0.63 substantially outperforming a similar network trained on the same set of examples from scratch.
* 8 pages, 3 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge