Chuanrui Wang
RealPDEBench: A Benchmark for Complex Physical Systems with Real-World Data
Jan 05, 2026Abstract:Predicting the evolution of complex physical systems remains a central problem in science and engineering. Despite rapid progress in scientific Machine Learning (ML) models, a critical bottleneck is the lack of expensive real-world data, resulting in most current models being trained and validated on simulated data. Beyond limiting the development and evaluation of scientific ML, this gap also hinders research into essential tasks such as sim-to-real transfer. We introduce RealPDEBench, the first benchmark for scientific ML that integrates real-world measurements with paired numerical simulations. RealPDEBench consists of five datasets, three tasks, eight metrics, and ten baselines. We first present five real-world measured datasets with paired simulated datasets across different complex physical systems. We further define three tasks, which allow comparisons between real-world and simulated data, and facilitate the development of methods to bridge the two. Moreover, we design eight evaluation metrics, spanning data-oriented and physics-oriented metrics, and finally benchmark ten representative baselines, including state-of-the-art models, pretrained PDE foundation models, and a traditional method. Experiments reveal significant discrepancies between simulated and real-world data, while showing that pretraining with simulated data consistently improves both accuracy and convergence. In this work, we hope to provide insights from real-world data, advancing scientific ML toward bridging the sim-to-real gap and real-world deployment. Our benchmark, datasets, and instructions are available at https://realpdebench.github.io/.
GGBall: Graph Generative Model on Poincaré Ball
Jun 08, 2025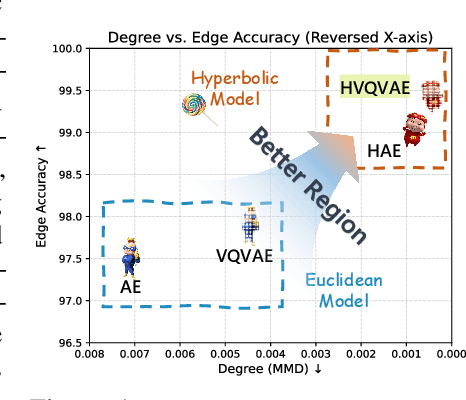
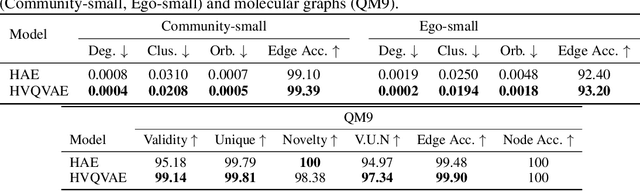
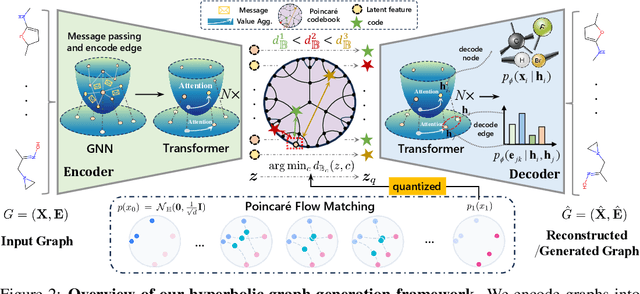
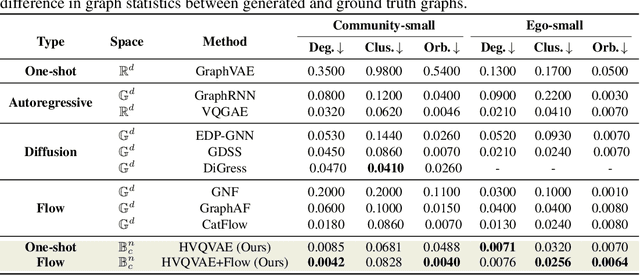
Abstract:Generating graphs with hierarchical structures remains a fundamental challenge due to the limitations of Euclidean geometry in capturing exponential complexity. Here we introduce \textbf{GGBall}, a novel hyperbolic framework for graph generation that integrates geometric inductive biases with modern generative paradigms. GGBall combines a Hyperbolic Vector-Quantized Autoencoder (HVQVAE) with a Riemannian flow matching prior defined via closed-form geodesics. This design enables flow-based priors to model complex latent distributions, while vector quantization helps preserve the curvature-aware structure of the hyperbolic space. We further develop a suite of hyperbolic GNN and Transformer layers that operate entirely within the manifold, ensuring stability and scalability. Empirically, our model reduces degree MMD by over 75\% on Community-Small and over 40\% on Ego-Small compared to state-of-the-art baselines, demonstrating an improved ability to preserve topological hierarchies. These results highlight the potential of hyperbolic geometry as a powerful foundation for the generative modeling of complex, structured, and hierarchical data domains. Our code is available at \href{https://github.com/AI4Science-WestlakeU/GGBall}{here}.
PDB-Struct: A Comprehensive Benchmark for Structure-based Protein Design
Nov 30, 2023



Abstract:Structure-based protein design has attracted increasing interest, with numerous methods being introduced in recent years. However, a universally accepted method for evaluation has not been established, since the wet-lab validation can be overly time-consuming for the development of new algorithms, and the $\textit{in silico}$ validation with recovery and perplexity metrics is efficient but may not precisely reflect true foldability. To address this gap, we introduce two novel metrics: refoldability-based metric, which leverages high-accuracy protein structure prediction models as a proxy for wet lab experiments, and stability-based metric, which assesses whether models can assign high likelihoods to experimentally stable proteins. We curate datasets from high-quality CATH protein data, high-throughput $\textit{de novo}$ designed proteins, and mega-scale experimental mutagenesis experiments, and in doing so, present the $\textbf{PDB-Struct}$ benchmark that evaluates both recent and previously uncompared protein design methods. Experimental results indicate that ByProt, ProteinMPNN, and ESM-IF perform exceptionally well on our benchmark, while ESM-Design and AF-Design fall short on the refoldability metric. We also show that while some methods exhibit high sequence recovery, they do not perform as well on our new benchmark. Our proposed benchmark paves the way for a fair and comprehensive evaluation of protein design methods in the future. Code is available at https://github.com/WANG-CR/PDB-Struct.
Protein Sequence and Structure Co-Design with Equivariant Translation
Oct 17, 2022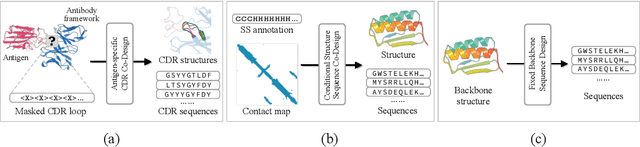
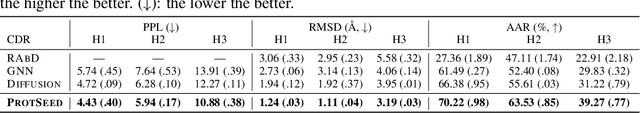
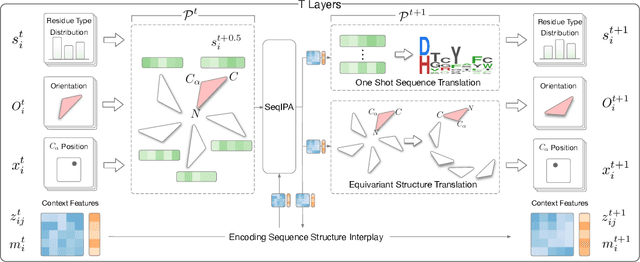
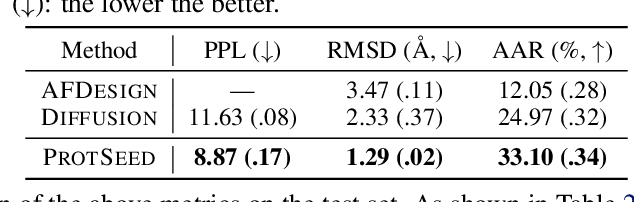
Abstract:Proteins are macromolecules that perform essential functions in all living organisms. Designing novel proteins with specific structures and desired functions has been a long-standing challenge in the field of bioengineering. Existing approaches generate both protein sequence and structure using either autoregressive models or diffusion models, both of which suffer from high inference costs. In this paper, we propose a new approach capable of protein sequence and structure co-design, which iteratively translates both protein sequence and structure into the desired state from random initialization, based on context features given a priori. Our model consists of a trigonometry-aware encoder that reasons geometrical constraints and interactions from context features, and a roto-translation equivariant decoder that translates protein sequence and structure interdependently. Notably, all protein amino acids are updated in one shot in each translation step, which significantly accelerates the inference process. Experimental results across multiple tasks show that our model outperforms previous state-of-the-art baselines by a large margin, and is able to design proteins of high fidelity as regards both sequence and structure, with running time orders of magnitude less than sampling-based methods.
Colossal-AI: A Unified Deep Learning System For Large-Scale Parallel Training
Oct 28, 2021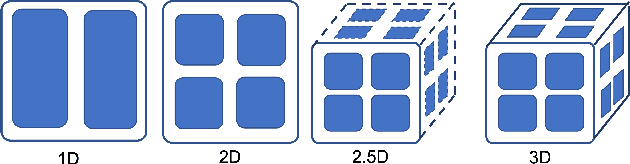
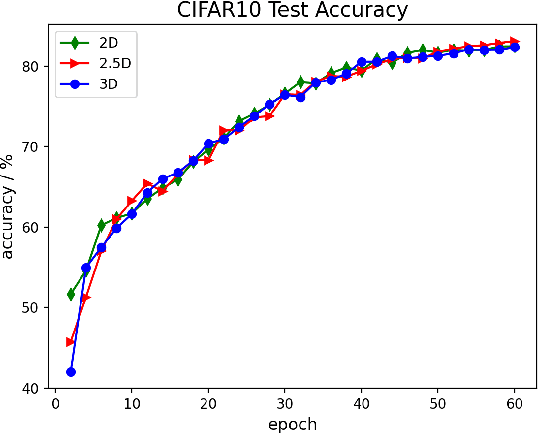
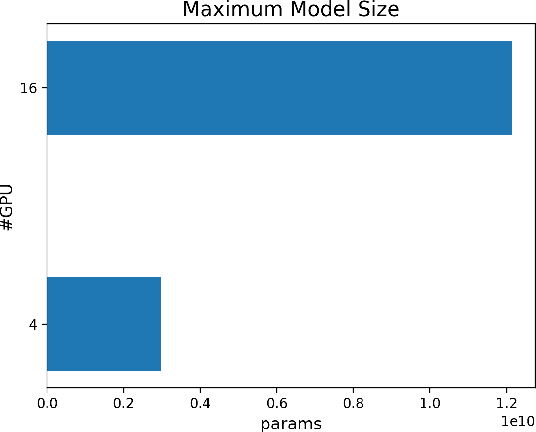
Abstract:The Transformer architecture has improved the performance of deep learning models in domains such as Computer Vision and Natural Language Processing. Together with better performance come larger model sizes. This imposes challenges to the memory wall of the current accelerator hardware such as GPU. It is never ideal to train large models such as Vision Transformer, BERT, and GPT on a single GPU or a single machine. There is an urgent demand to train models in a distributed environment. However, distributed training, especially model parallelism, often requires domain expertise in computer systems and architecture. It remains a challenge for AI researchers to implement complex distributed training solutions for their models. In this paper, we introduce Colossal-AI, which is a unified parallel training system designed to seamlessly integrate different paradigms of parallelization techniques including data parallelism, pipeline parallelism, multiple tensor parallelism, and sequence parallelism. Colossal-AI aims to support the AI community to write distributed models in the same way as how they write models normally. This allows them to focus on developing the model architecture and separates the concerns of distributed training from the development process. The documentations can be found at https://www.colossalai.org and the source code can be found at https://github.com/hpcaitech/ColossalAI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge