Christoph Angerer
Object-Level Targeted Selection via Deep Template Matching
Jul 05, 2022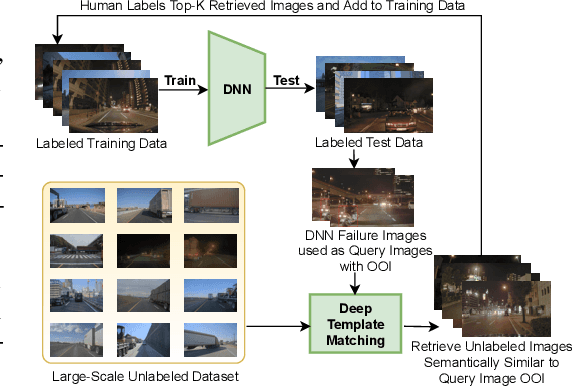
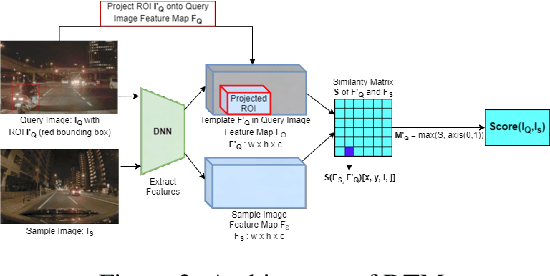
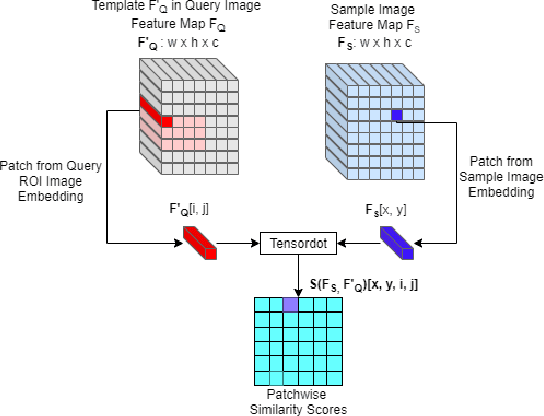
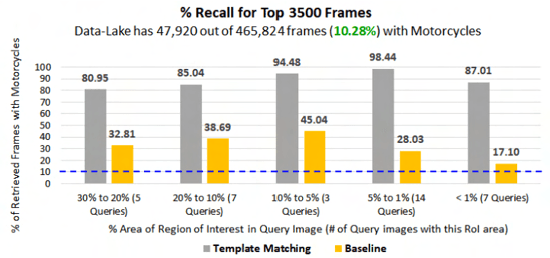
Abstract:Retrieving images with objects that are semantically similar to objects of interest (OOI) in a query image has many practical use cases. A few examples include fixing failures like false negatives/positives of a learned model or mitigating class imbalance in a dataset. The targeted selection task requires finding the relevant data from a large-scale pool of unlabeled data. Manual mining at this scale is infeasible. Further, the OOI are often small and occupy less than 1% of image area, are occluded, and co-exist with many semantically different objects in cluttered scenes. Existing semantic image retrieval methods often focus on mining for larger sized geographical landmarks, and/or require extra labeled data, such as images/image-pairs with similar objects, for mining images with generic objects. We propose a fast and robust template matching algorithm in the DNN feature space, that retrieves semantically similar images at the object-level from a large unlabeled pool of data. We project the region(s) around the OOI in the query image to the DNN feature space for use as the template. This enables our method to focus on the semantics of the OOI without requiring extra labeled data. In the context of autonomous driving, we evaluate our system for targeted selection by using failure cases of object detectors as OOI. We demonstrate its efficacy on a large unlabeled dataset with 2.2M images and show high recall in mining for images with small-sized OOI. We compare our method against a well-known semantic image retrieval method, which also does not require extra labeled data. Lastly, we show that our method is flexible and retrieves images with one or more semantically different co-occurring OOI seamlessly.
CodeTrans: Towards Cracking the Language of Silicone's Code Through Self-Supervised Deep Learning and High Performance Computing
Apr 06, 2021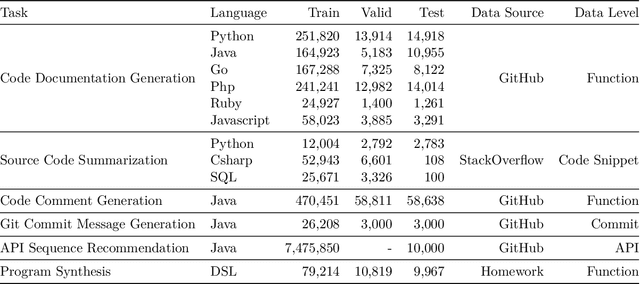
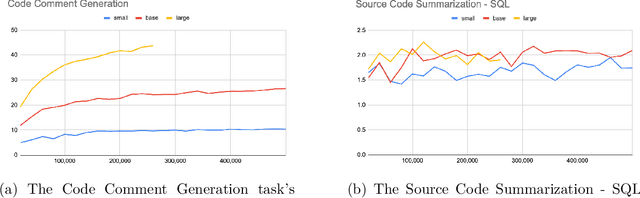
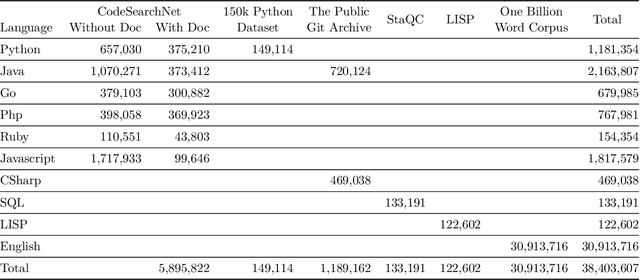
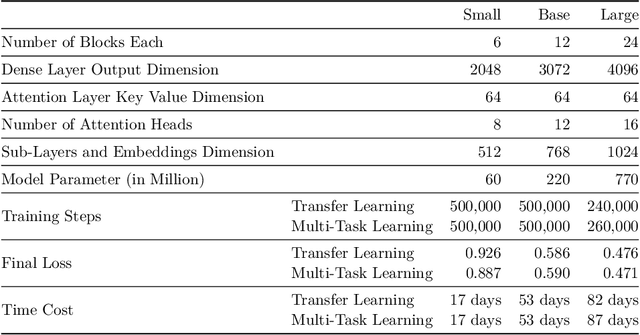
Abstract:Currently, a growing number of mature natural language processing applications make people's life more convenient. Such applications are built by source code - the language in software engineering. However, the applications for understanding source code language to ease the software engineering process are under-researched. Simultaneously, the transformer model, especially its combination with transfer learning, has been proven to be a powerful technique for natural language processing tasks. These breakthroughs point out a promising direction for process source code and crack software engineering tasks. This paper describes CodeTrans - an encoder-decoder transformer model for tasks in the software engineering domain, that explores the effectiveness of encoder-decoder transformer models for six software engineering tasks, including thirteen sub-tasks. Moreover, we have investigated the effect of different training strategies, including single-task learning, transfer learning, multi-task learning, and multi-task learning with fine-tuning. CodeTrans outperforms the state-of-the-art models on all the tasks. To expedite future works in the software engineering domain, we have published our pre-trained models of CodeTrans. https://github.com/agemagician/CodeTrans
ProtTrans: Towards Cracking the Language of Life's Code Through Self-Supervised Deep Learning and High Performance Computing
Jul 20, 2020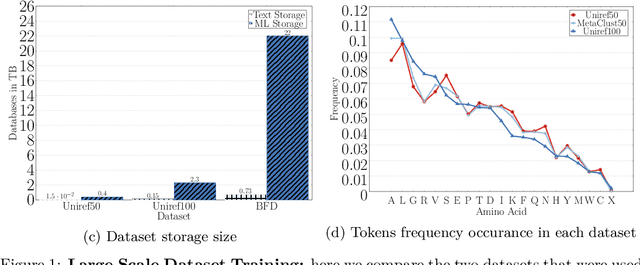
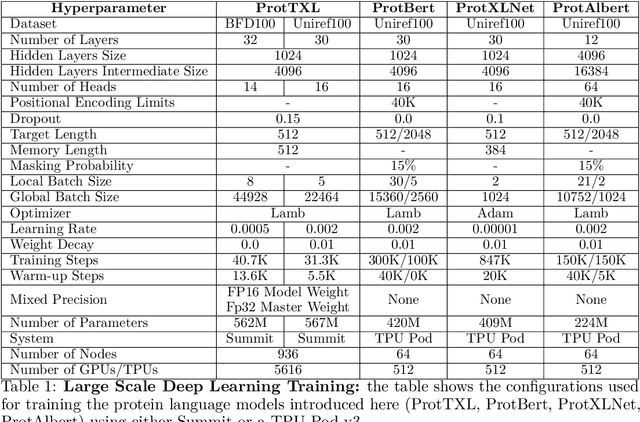
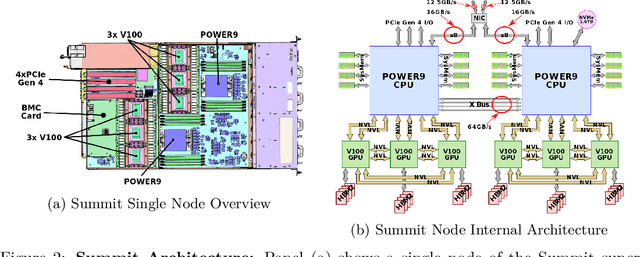
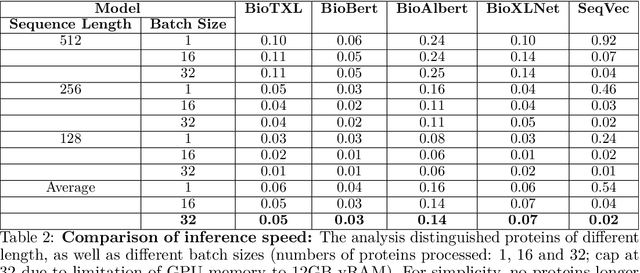
Abstract:Computational biology and bioinformatics provide vast data gold-mines from protein sequences, ideal for Language Models (LMs) taken from Natural Language Processing (NLP). These LMs reach for new prediction frontiers at low inference costs. Here, we trained two auto-regressive language models (Transformer-XL, XLNet) and two auto-encoder models (Bert, Albert) on data from UniRef and BFD containing up to 393 billion amino acids (words) from 2.1 billion protein sequences (22- and 112-times the entire English Wikipedia). The LMs were trained on the Summit supercomputer at Oak Ridge National Laboratory (ORNL), using 936 nodes (total 5616 GPUs) and one TPU Pod (V3-512 or V3-1024). We validated the advantage of up-scaling LMs to larger models supported by bigger data by predicting secondary structure (3-states: Q3=76-84, 8-states: Q8=65-73), sub-cellular localization for 10 cellular compartments (Q10=74) and whether a protein is membrane-bound or water-soluble (Q2=89). Dimensionality reduction revealed that the LM-embeddings from unlabeled data (only protein sequences) captured important biophysical properties governing protein shape. This implied learning some of the grammar of the language of life realized in protein sequences. The successful up-scaling of protein LMs through HPC to larger data sets slightly reduced the gap between models trained on evolutionary information and LMs. The official GitHub repository: https://github.com/agemagician/ProtTrans
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge