Tamas Feher
CAGRA: Highly Parallel Graph Construction and Approximate Nearest Neighbor Search for GPUs
Aug 29, 2023



Abstract:Approximate Nearest Neighbor Search (ANNS) plays a critical role in various disciplines spanning data mining and artificial intelligence, from information retrieval and computer vision to natural language processing and recommender systems. Data volumes have soared in recent years and the computational cost of an exhaustive exact nearest neighbor search is often prohibitive, necessitating the adoption of approximate techniques. The balanced performance and recall of graph-based approaches have more recently garnered significant attention in ANNS algorithms, however, only a few studies have explored harnessing the power of GPUs and multi-core processors despite the widespread use of massively parallel and general-purpose computing. To bridge this gap, we introduce a novel parallel computing hardware-based proximity graph and search algorithm. By leveraging the high-performance capabilities of modern hardware, our approach achieves remarkable efficiency gains. In particular, our method surpasses existing CPU and GPU-based methods in constructing the proximity graph, demonstrating higher throughput in both large- and small-batch searches while maintaining compatible accuracy. In graph construction time, our method, CAGRA, is 2.2~27x faster than HNSW, which is one of the CPU SOTA implementations. In large-batch query throughput in the 90% to 95% recall range, our method is 33~77x faster than HNSW, and is 3.8~8.8x faster than the SOTA implementations for GPU. For a single query, our method is 3.4~53x faster than HNSW at 95% recall.
CodeTrans: Towards Cracking the Language of Silicone's Code Through Self-Supervised Deep Learning and High Performance Computing
Apr 06, 2021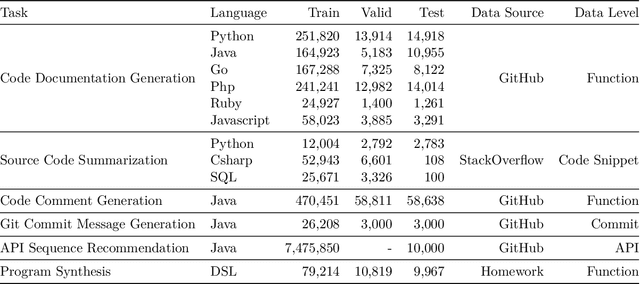
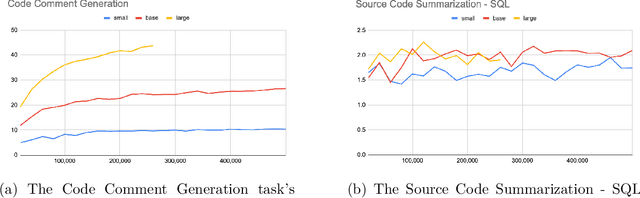
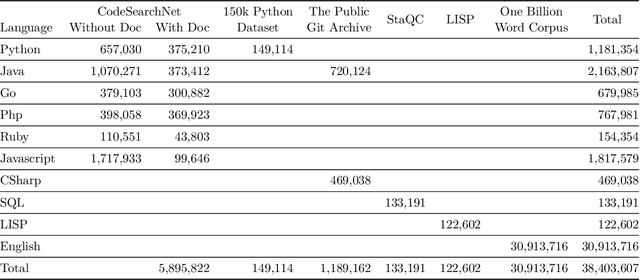
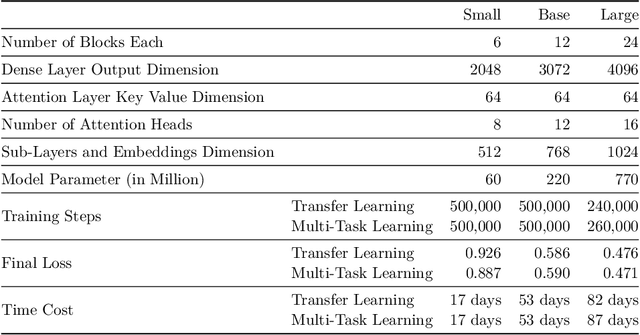
Abstract:Currently, a growing number of mature natural language processing applications make people's life more convenient. Such applications are built by source code - the language in software engineering. However, the applications for understanding source code language to ease the software engineering process are under-researched. Simultaneously, the transformer model, especially its combination with transfer learning, has been proven to be a powerful technique for natural language processing tasks. These breakthroughs point out a promising direction for process source code and crack software engineering tasks. This paper describes CodeTrans - an encoder-decoder transformer model for tasks in the software engineering domain, that explores the effectiveness of encoder-decoder transformer models for six software engineering tasks, including thirteen sub-tasks. Moreover, we have investigated the effect of different training strategies, including single-task learning, transfer learning, multi-task learning, and multi-task learning with fine-tuning. CodeTrans outperforms the state-of-the-art models on all the tasks. To expedite future works in the software engineering domain, we have published our pre-trained models of CodeTrans. https://github.com/agemagician/CodeTrans
ProtTrans: Towards Cracking the Language of Life's Code Through Self-Supervised Deep Learning and High Performance Computing
Jul 20, 2020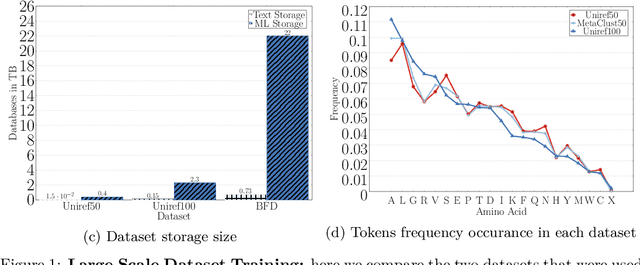
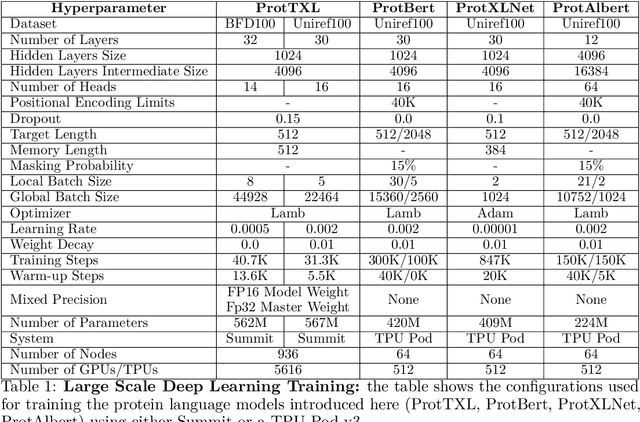
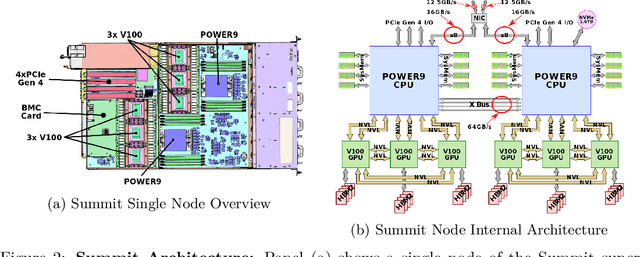
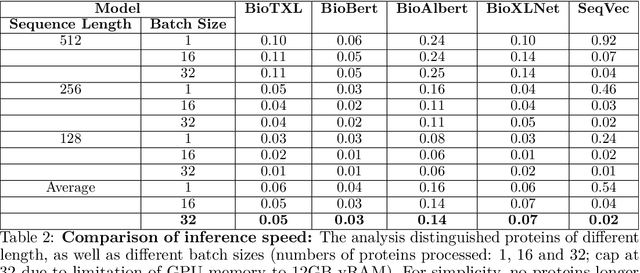
Abstract:Computational biology and bioinformatics provide vast data gold-mines from protein sequences, ideal for Language Models (LMs) taken from Natural Language Processing (NLP). These LMs reach for new prediction frontiers at low inference costs. Here, we trained two auto-regressive language models (Transformer-XL, XLNet) and two auto-encoder models (Bert, Albert) on data from UniRef and BFD containing up to 393 billion amino acids (words) from 2.1 billion protein sequences (22- and 112-times the entire English Wikipedia). The LMs were trained on the Summit supercomputer at Oak Ridge National Laboratory (ORNL), using 936 nodes (total 5616 GPUs) and one TPU Pod (V3-512 or V3-1024). We validated the advantage of up-scaling LMs to larger models supported by bigger data by predicting secondary structure (3-states: Q3=76-84, 8-states: Q8=65-73), sub-cellular localization for 10 cellular compartments (Q10=74) and whether a protein is membrane-bound or water-soluble (Q2=89). Dimensionality reduction revealed that the LM-embeddings from unlabeled data (only protein sequences) captured important biophysical properties governing protein shape. This implied learning some of the grammar of the language of life realized in protein sequences. The successful up-scaling of protein LMs through HPC to larger data sets slightly reduced the gap between models trained on evolutionary information and LMs. The official GitHub repository: https://github.com/agemagician/ProtTrans
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge