Bella Specktor Fadida
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Contour Dice loss for structures with Fuzzy and Complex Boundaries in Fetal MRI
Sep 25, 2022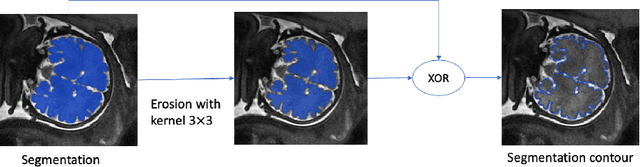
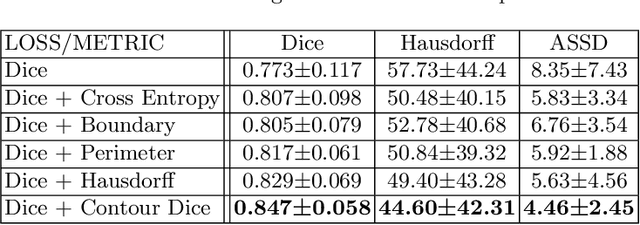
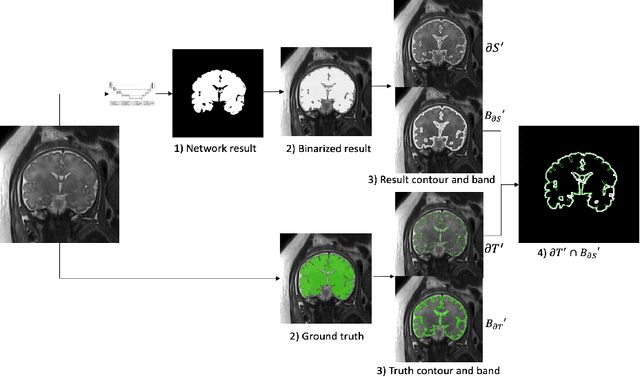
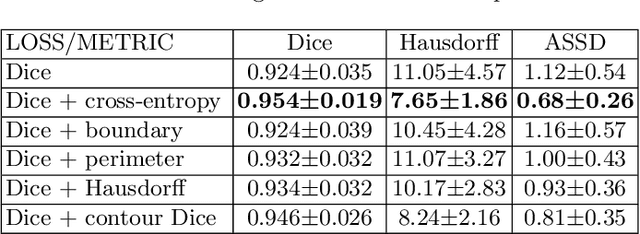
Abstract:Volumetric measurements of fetal structures in MRI are time consuming and error prone and therefore require automatic segmentation. Placenta segmentation and accurate fetal brain segmentation for gyrification assessment are particularly challenging because of the placenta fuzzy boundaries and the fetal brain cortex complex foldings. In this paper, we study the use of the Contour Dice loss for both problems and compare it to other boundary losses and to the combined Dice and Cross-Entropy loss. The loss is computed efficiently for each slice via erosion, dilation and XOR operators. We describe a new formulation of the loss akin to the Contour Dice metric. The combination of the Dice loss and the Contour Dice yielded the best performance for placenta segmentation. For fetal brain segmentation, the best performing loss was the combined Dice with Cross-Entropy loss followed by the Dice with Contour Dice loss, which performed better than other boundary losses.
Partial annotations for the segmentation of large structures with low annotation cost
Sep 25, 2022Abstract:Deep learning methods have been shown to be effective for the automatic segmentation of structures and pathologies in medical imaging. However, they require large annotated datasets, whose manual segmentation is a tedious and time-consuming task, especially for large structures. We present a new method of partial annotations that uses a small set of consecutive annotated slices from each scan with an annotation effort that is equal to that of only few annotated cases. The training with partial annotations is performed by using only annotated blocks, incorporating information about slices outside the structure of interest and modifying a batch loss function to consider only the annotated slices. To facilitate training in a low data regime, we use a two-step optimization process. We tested the method with the popular soft Dice loss for the fetal body segmentation task in two MRI sequences, TRUFI and FIESTA, and compared full annotation regime to partial annotations with a similar annotation effort. For TRUFI data, the use of partial annotations yielded slightly better performance on average compared to full annotations with an increase in Dice score from 0.936 to 0.942, and a substantial decrease in Standard Deviations (STD) of Dice score by 22% and Average Symmetric Surface Distance (ASSD) by 15%. For the FIESTA sequence, partial annotations also yielded a decrease in STD of the Dice score and ASSD metrics by 27.5% and 33% respectively for in-distribution data, and a substantial improvement also in average performance on out-of-distribution data, increasing Dice score from 0.84 to 0.9 and decreasing ASSD from 7.46 to 4.01 mm. The two-step optimization process was helpful for partial annotations for both in-distribution and out-of-distribution data. The partial annotations method with the two-step optimizer is therefore recommended to improve segmentation performance under low data regime.
* 10 pages, 4 figures
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022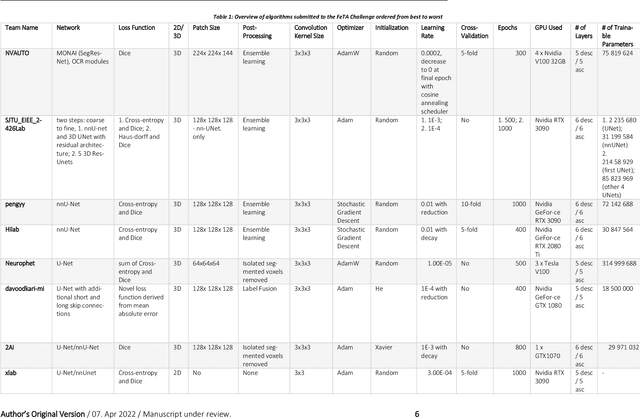
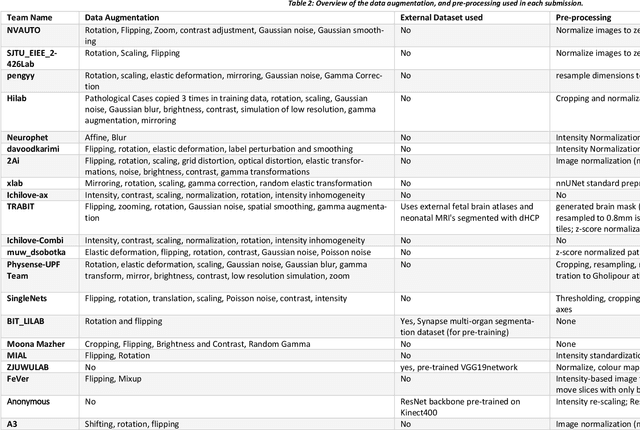
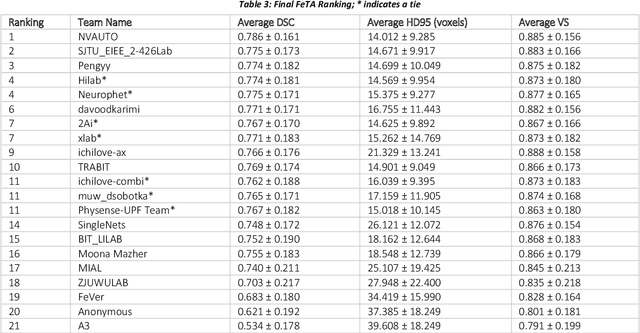
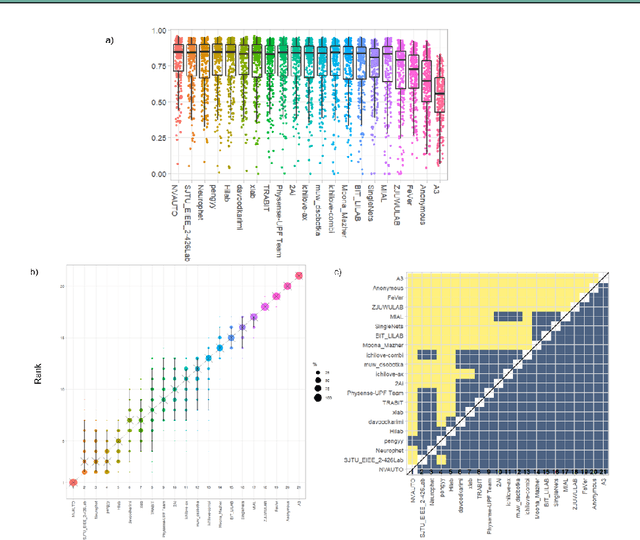
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge