Antonio Di Ieva
Towards a Multimodal MRI-Based Foundation Model for Multi-Level Feature Exploration in Segmentation, Molecular Subtyping, and Grading of Glioma
Mar 10, 2025Abstract:Accurate, noninvasive glioma characterization is crucial for effective clinical management. Traditional methods, dependent on invasive tissue sampling, often fail to capture the spatial heterogeneity of the tumor. While deep learning has improved segmentation and molecular profiling, few approaches simultaneously integrate tumor morphology and molecular features. Foundation deep learning models, which learn robust, task-agnostic representations from large-scale datasets, hold great promise but remain underutilized in glioma imaging biomarkers. We propose the Multi-Task SWIN-UNETR (MTS-UNET) model, a novel foundation-based framework built on the BrainSegFounder model, pretrained on large-scale neuroimaging data. MTS-UNET simultaneously performs glioma segmentation, histological grading, and molecular subtyping (IDH mutation and 1p/19q co-deletion). It incorporates two key modules: Tumor-Aware Feature Encoding (TAFE) for multi-scale, tumor-focused feature extraction and Cross-Modality Differential (CMD) for highlighting subtle T2-FLAIR mismatch signals associated with IDH mutation. The model was trained and validated on a diverse, multi-center cohort of 2,249 glioma patients from seven public datasets. MTS-UNET achieved a mean Dice score of 84% for segmentation, along with AUCs of 90.58% for IDH mutation, 69.22% for 1p/19q co-deletion prediction, and 87.54% for grading, significantly outperforming baseline models (p<=0.05). Ablation studies validated the essential contributions of the TAFE and CMD modules and demonstrated the robustness of the framework. The foundation-based MTS-UNET model effectively integrates tumor segmentation with multi-level classification, exhibiting strong generalizability across diverse MRI datasets. This framework shows significant potential for advancing noninvasive, personalized glioma management by improving predictive accuracy and interpretability.
Diagnostic Performance of Deep Learning for Predicting Gliomas' IDH and 1p/19q Status in MRI: A Systematic Review and Meta-Analysis
Oct 28, 2024Abstract:Gliomas, the most common primary brain tumors, show high heterogeneity in histological and molecular characteristics. Accurate molecular profiling, like isocitrate dehydrogenase (IDH) mutation and 1p/19q codeletion, is critical for diagnosis, treatment, and prognosis. This review evaluates MRI-based deep learning (DL) models' efficacy in predicting these biomarkers. Following PRISMA guidelines, we systematically searched major databases (PubMed, Scopus, Ovid, and Web of Science) up to February 2024, screening studies that utilized DL to predict IDH and 1p/19q codeletion status from MRI data of glioma patients. We assessed the quality and risk of bias using the radiomics quality score and QUADAS-2 tool. Our meta-analysis used a bivariate model to compute pooled sensitivity, specificity, and meta-regression to assess inter-study heterogeneity. Of the 565 articles, 57 were selected for qualitative synthesis, and 52 underwent meta-analysis. The pooled estimates showed high diagnostic performance, with validation sensitivity, specificity, and area under the curve (AUC) of 0.84 [prediction interval (PI): 0.67-0.93, I2=51.10%, p < 0.05], 0.87 [PI: 0.49-0.98, I2=82.30%, p < 0.05], and 0.89 for IDH prediction, and 0.76 [PI: 0.28-0.96, I2=77.60%, p < 0.05], 0.85 [PI: 0.49-0.97, I2=80.30%, p < 0.05], and 0.90 for 1p/19q prediction, respectively. Meta-regression analyses revealed significant heterogeneity influenced by glioma grade, data source, inclusion of non-radiomics data, MRI sequences, segmentation and feature extraction methods, and validation techniques. DL models demonstrate strong potential in predicting molecular biomarkers from MRI scans, with significant variability influenced by technical and clinical factors. Thorough external validation is necessary to increase clinical utility.
When Eye-Tracking Meets Machine Learning: A Systematic Review on Applications in Medical Image Analysis
Mar 12, 2024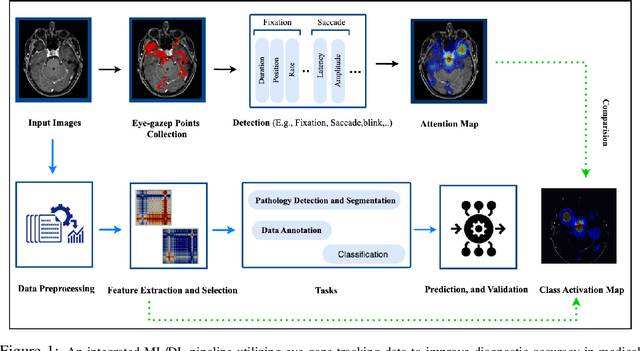
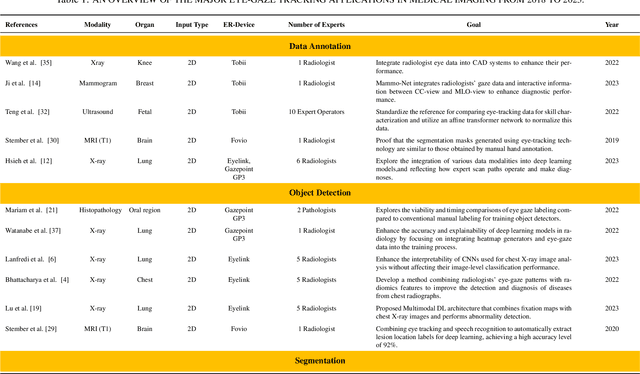
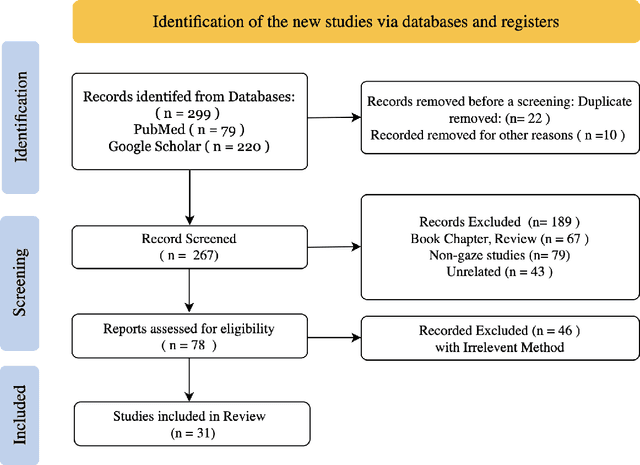
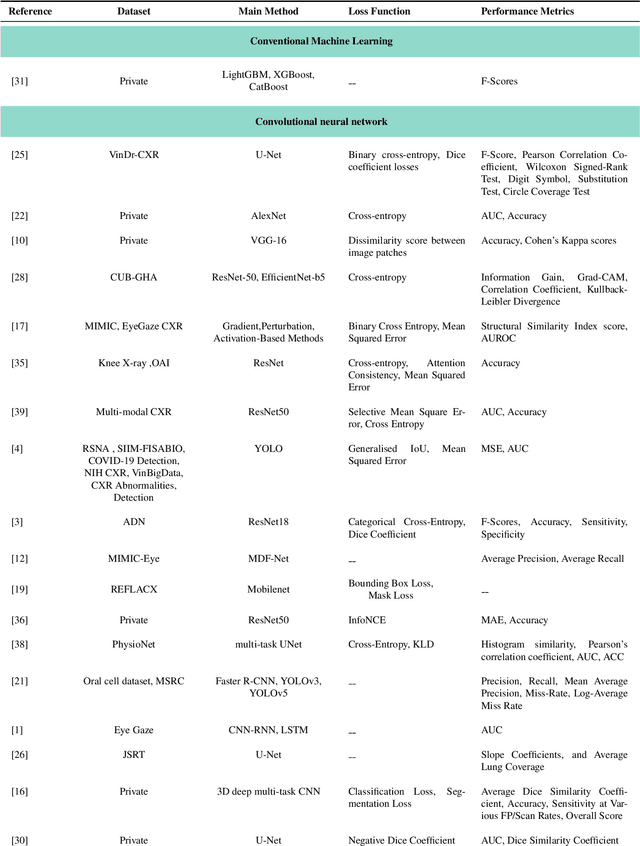
Abstract:Eye-gaze tracking research offers significant promise in enhancing various healthcare-related tasks, above all in medical image analysis and interpretation. Eye tracking, a technology that monitors and records the movement of the eyes, provides valuable insights into human visual attention patterns. This technology can transform how healthcare professionals and medical specialists engage with and analyze diagnostic images, offering a more insightful and efficient approach to medical diagnostics. Hence, extracting meaningful features and insights from medical images by leveraging eye-gaze data improves our understanding of how radiologists and other medical experts monitor, interpret, and understand images for diagnostic purposes. Eye-tracking data, with intricate human visual attention patterns embedded, provides a bridge to integrating artificial intelligence (AI) development and human cognition. This integration allows novel methods to incorporate domain knowledge into machine learning (ML) and deep learning (DL) approaches to enhance their alignment with human-like perception and decision-making. Moreover, extensive collections of eye-tracking data have also enabled novel ML/DL methods to analyze human visual patterns, paving the way to a better understanding of human vision, attention, and cognition. This systematic review investigates eye-gaze tracking applications and methodologies for enhancing ML/DL algorithms for medical image analysis in depth.
TPMIL: Trainable Prototype Enhanced Multiple Instance Learning for Whole Slide Image Classification
May 01, 2023Abstract:Digital pathology based on whole slide images (WSIs) plays a key role in cancer diagnosis and clinical practice. Due to the high resolution of the WSI and the unavailability of patch-level annotations, WSI classification is usually formulated as a weakly supervised problem, which relies on multiple instance learning (MIL) based on patches of a WSI. In this paper, we aim to learn an optimal patch-level feature space by integrating prototype learning with MIL. To this end, we develop a Trainable Prototype enhanced deep MIL (TPMIL) framework for weakly supervised WSI classification. In contrast to the conventional methods which rely on a certain number of selected patches for feature space refinement, we softly cluster all the instances by allocating them to their corresponding prototypes. Additionally, our method is able to reveal the correlations between different tumor subtypes through distances between corresponding trained prototypes. More importantly, TPMIL also enables to provide a more accurate interpretability based on the distance of the instances from the trained prototypes which serves as an alternative to the conventional attention score-based interpretability. We test our method on two WSI datasets and it achieves a new SOTA. GitHub repository: https://github.com/LitaoYang-Jet/TPMIL
Domain-Specific Pretraining Improves Confidence in Whole Slide Image Classification
Feb 20, 2023Abstract:Whole Slide Images (WSIs) or histopathology images are used in digital pathology. WSIs pose great challenges to deep learning models for clinical diagnosis, owing to their size and lack of pixel-level annotations. With the recent advancements in computational pathology, newer multiple-instance learning-based models have been proposed. Multiple-instance learning for WSIs necessitates creating patches and uses the encoding of these patches for diagnosis. These models use generic pre-trained models (ResNet-50 pre-trained on ImageNet) for patch encoding. The recently proposed KimiaNet, a DenseNet121 model pre-trained on TCGA slides, is a domain-specific pre-trained model. This paper shows the effect of domain-specific pre-training on WSI classification. To investigate the impact of domain-specific pre-training, we considered the current state-of-the-art multiple-instance learning models, 1) CLAM, an attention-based model, and 2) TransMIL, a self-attention-based model, and evaluated the models' confidence and predictive performance in detecting primary brain tumors - gliomas. Domain-specific pre-training improves the confidence of the models and also achieves a new state-of-the-art performance of WSI-based glioma subtype classification, showing a high clinical applicability in assisting glioma diagnosis.
Impact of Spherical Coordinates Transformation Pre-processing in Deep Convolution Neural Networks for Brain Tumor Segmentation and Survival Prediction
Oct 27, 2020


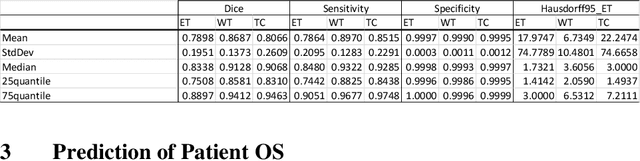
Abstract:Pre-processing and Data Augmentation play an important role in Deep Convolutional Neural Networks (DCNN). Whereby several methods aim for standardization and augmentation of the dataset, we here propose a novel method aimed to feed DCNN with spherical space transformed input data that could better facilitate feature learning compared to standard Cartesian space images and volumes. In this work, the spherical coordinates transformation has been applied as a preprocessing method that, used in conjunction with normal MRI volumes, improves the accuracy of brain tumor segmentation and patient overall survival (OS) prediction on Brain Tumor Segmentation (BraTS) Challenge 2020 dataset. The LesionEncoder framework has been then applied to automatically extract features from DCNN models, achieving 0.586 accuracy of OS prediction on the validation data set, which is one of the best results according to BraTS 2020 leaderboard.
Spherical coordinates transformation pre-processing in Deep Convolution Neural Networks for brain tumor segmentation in MRI
Aug 17, 2020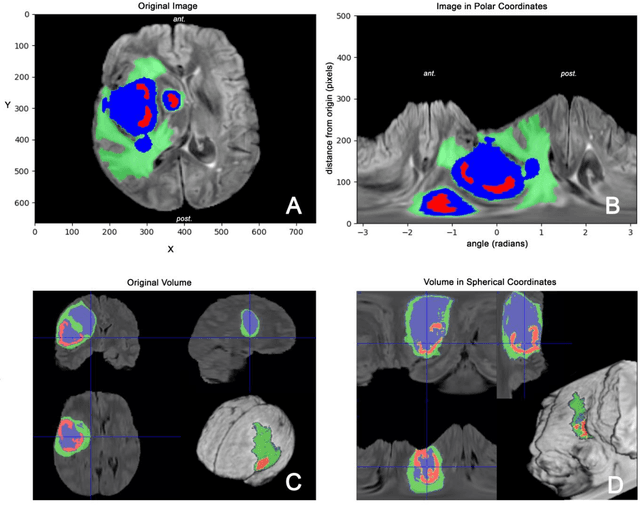



Abstract:Magnetic Resonance Imaging (MRI) is used in everyday clinical practice to assess brain tumors. Several automatic or semi-automatic segmentation algorithms have been introduced to segment brain tumors and achieve an expert-like accuracy. Deep Convolutional Neural Networks (DCNN) have recently shown very promising results, however, DCNN models are still far from achieving clinically meaningful results mainly because of the lack of generalization of the models. DCNN models need large annotated datasets to achieve good performance. Models are often optimized on the domain dataset on which they have been trained, and then fail the task when the same model is applied to different datasets from different institutions. One of the reasons is due to the lack of data standardization to adjust for different models and MR machines. In this work, a 3D Spherical coordinates transform during the pre-processing phase has been hypothesized to improve DCNN models' accuracy and to allow more generalizable results even when the model is trained on small and heterogeneous datasets and translated into different domains. Indeed, the spherical coordinate system avoids several standardization issues since it works independently of resolution and imaging settings. Both Cartesian and spherical volumes were evaluated in two DCNN models with the same network structure using the BraTS 2019 dataset. The model trained on spherical transform pre-processed inputs resulted in superior performance over the Cartesian-input trained model on predicting gliomas' segmentation on tumor core and enhancing tumor classes (increase of 0.011 and 0.014 respectively on the validation dataset), achieving a further improvement in accuracy by merging the two models together. Furthermore, the spherical transform is not resolution-dependent and achieve same results on different input resolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge