Spherical coordinates transformation pre-processing in Deep Convolution Neural Networks for brain tumor segmentation in MRI
Paper and Code
Aug 17, 2020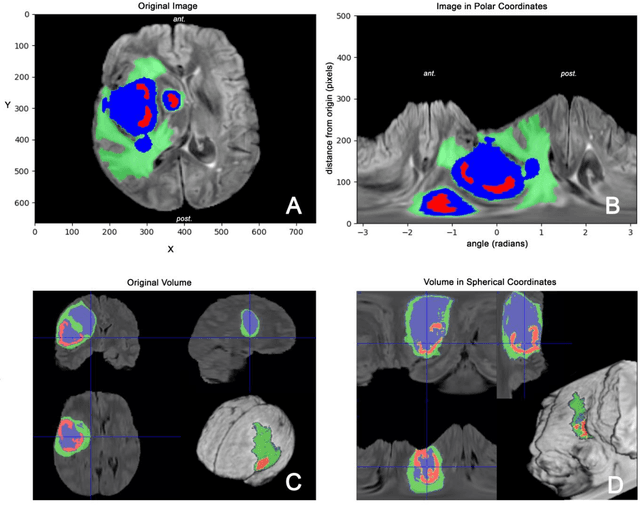



Magnetic Resonance Imaging (MRI) is used in everyday clinical practice to assess brain tumors. Several automatic or semi-automatic segmentation algorithms have been introduced to segment brain tumors and achieve an expert-like accuracy. Deep Convolutional Neural Networks (DCNN) have recently shown very promising results, however, DCNN models are still far from achieving clinically meaningful results mainly because of the lack of generalization of the models. DCNN models need large annotated datasets to achieve good performance. Models are often optimized on the domain dataset on which they have been trained, and then fail the task when the same model is applied to different datasets from different institutions. One of the reasons is due to the lack of data standardization to adjust for different models and MR machines. In this work, a 3D Spherical coordinates transform during the pre-processing phase has been hypothesized to improve DCNN models' accuracy and to allow more generalizable results even when the model is trained on small and heterogeneous datasets and translated into different domains. Indeed, the spherical coordinate system avoids several standardization issues since it works independently of resolution and imaging settings. Both Cartesian and spherical volumes were evaluated in two DCNN models with the same network structure using the BraTS 2019 dataset. The model trained on spherical transform pre-processed inputs resulted in superior performance over the Cartesian-input trained model on predicting gliomas' segmentation on tumor core and enhancing tumor classes (increase of 0.011 and 0.014 respectively on the validation dataset), achieving a further improvement in accuracy by merging the two models together. Furthermore, the spherical transform is not resolution-dependent and achieve same results on different input resolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge