António Cunha
Advancing Image-Based Grapevine Variety Classification with a New Benchmark and Evaluation of Masked Autoencoders
Jun 16, 2025Abstract:Grapevine varieties are essential for the economies of many wine-producing countries, influencing the production of wine, juice, and the consumption of fruits and leaves. Traditional identification methods, such as ampelography and molecular analysis, have limitations: ampelography depends on expert knowledge and is inherently subjective, while molecular methods are costly and time-intensive. To address these limitations, recent studies have applied deep learning (DL) models to classify grapevine varieties using image data. However, due to the small dataset sizes, these methods often depend on transfer learning from datasets from other domains, e.g., ImageNet1K (IN1K), which can lead to performance degradation due to domain shift and supervision collapse. In this context, self-supervised learning (SSL) methods can be a good tool to avoid this performance degradation, since they can learn directly from data, without external labels. This study presents an evaluation of Masked Autoencoders (MAEs) for identifying grapevine varieties based on field-acquired images. The main contributions of this study include two benchmarks comprising 43 grapevine varieties collected across different seasons, an analysis of MAE's application in the agricultural context, and a performance comparison of trained models across seasons. Our results show that a ViT-B/16 model pre-trained with MAE and the unlabeled dataset achieved an F1 score of 0.7956, outperforming all other models. Additionally, we observed that pre-trained models benefit from long pre-training, perform well under low-data training regime, and that simple data augmentation methods are more effective than complex ones. The study also found that the mask ratio in MAE impacts performance only marginally.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
LNDb: A Lung Nodule Database on Computed Tomography
Dec 19, 2019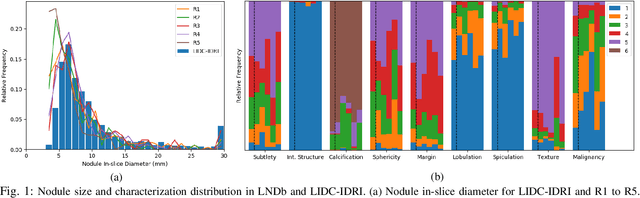
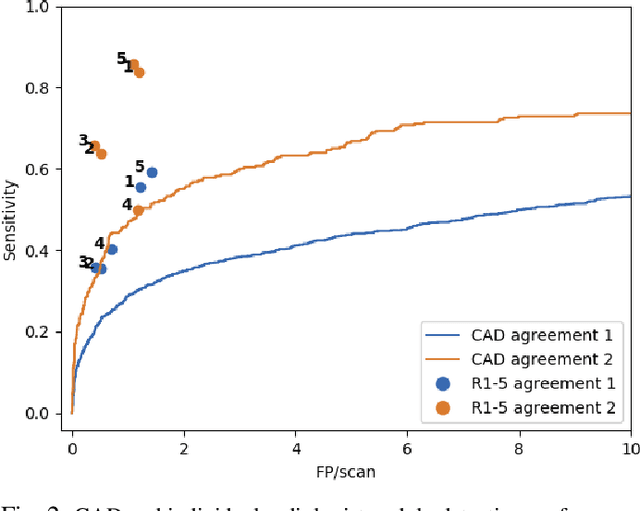
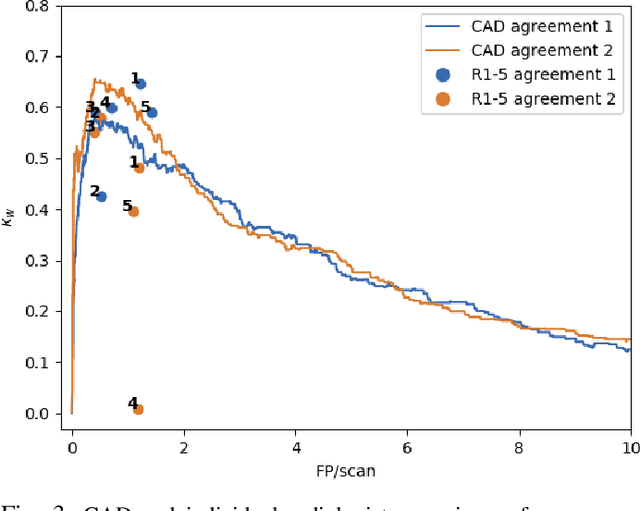
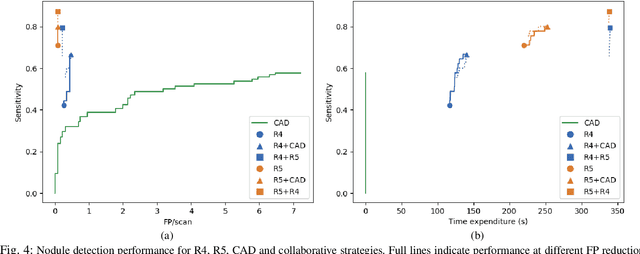
Abstract:Lung cancer is the deadliest type of cancer worldwide and late detection is the major factor for the low survival rate of patients. Low dose computed tomography has been suggested as a potential screening tool but manual screening is costly, time-consuming and prone to variability. This has fueled the development of automatic methods for the detection, segmentation and characterisation of pulmonary nodules but its application to clinical routine is challenging. In this study, a new database for the development and testing of pulmonary nodule computer-aided strategies is presented which intends to complement current databases by giving additional focus to radiologist variability and local clinical reality. State-of-the-art nodule detection, segmentation and characterization methods are tested and compared to manual annotations as well as collaborative strategies combining multiple radiologists and radiologists and computer-aided systems. It is shown that state-of-the-art methodologies can determine a patient's follow-up recommendation as accurately as a radiologist, though the nodule detection method used shows decreased performance in this database.
Did you miss it? Automatic lung nodule detection combined with gaze information improves radiologists' screening performance
Oct 09, 2019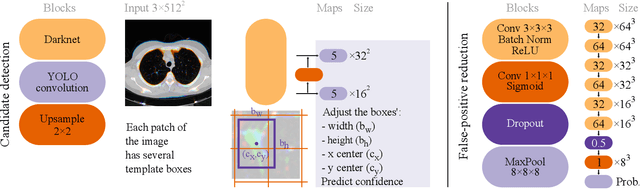
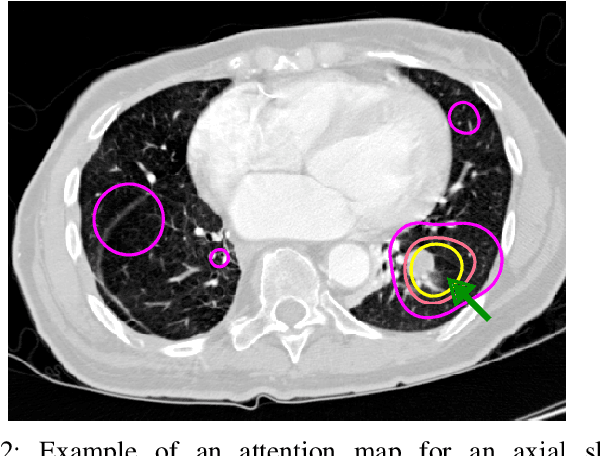
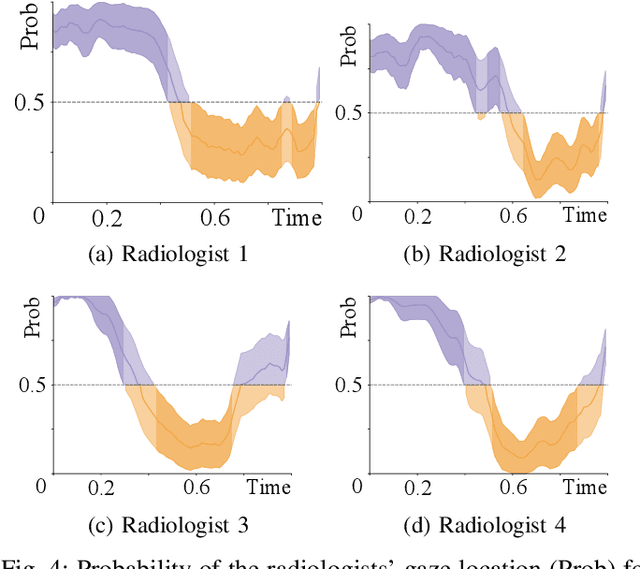
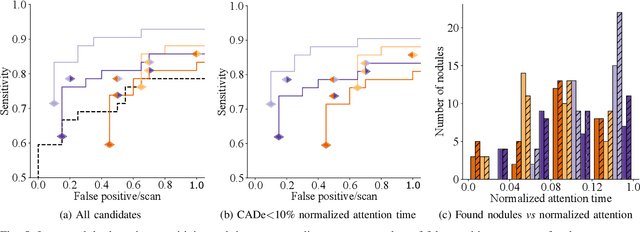
Abstract:Early diagnosis of lung cancer via computed tomography can significantly reduce the morbidity and mortality rates associated with the pathology. However, search lung nodules is a high complexity task, which affects the success of screening programs. Whilst computer-aided detection systems can be used as second observers, they may bias radiologists and introduce significant time overheads. With this in mind, this study assesses the potential of using gaze information for integrating automatic detection systems in the clinical practice. For that purpose, 4 radiologists were asked to annotate 20 scans from a public dataset while being monitored by an eye tracker device and an automatic lung nodule detection system was developed. Our results show that radiologists follow a similar search routine and tend to have lower fixation periods in regions where finding errors occur. The overall detection sensitivity of the specialists was 0.67$\pm$0.07, whereas the system achieved 0.69. Combining the annotations of one radiologist with the automatic system significantly improves the detection performance to similar levels of two annotators. Likewise, combining the findings of radiologist with the detection algorithm only for low fixation regions still significantly improves the detection sensitivity without increasing the number of false-positives. The combination of the automatic system with the gaze information allows to mitigate possible errors of the radiologist without some of the issues usually associated with automatic detection system.
iW-Net: an automatic and minimalistic interactive lung nodule segmentation deep network
Nov 30, 2018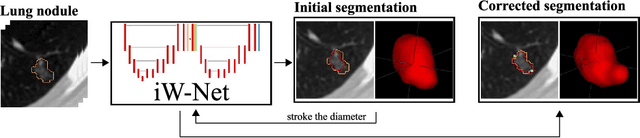
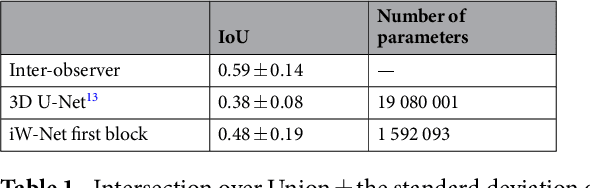
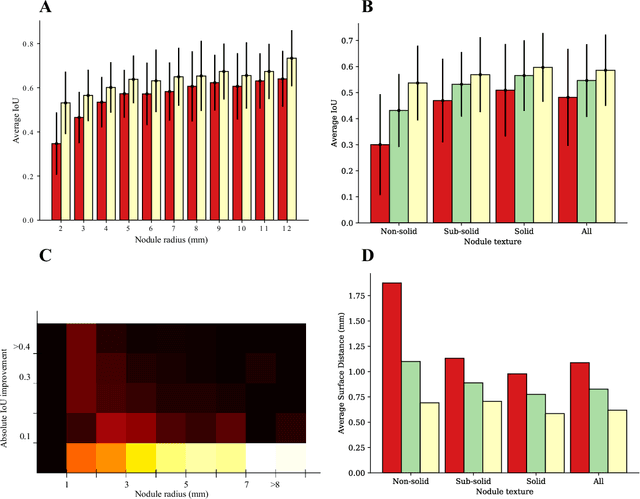
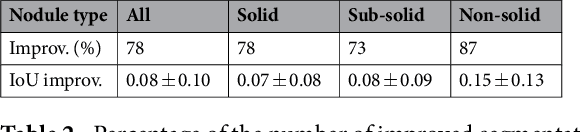
Abstract:We propose iW-Net, a deep learning model that allows for both automatic and interactive segmentation of lung nodules in computed tomography images. iW-Net is composed of two blocks: the first one provides an automatic segmentation and the second one allows to correct it by analyzing 2 points introduced by the user in the nodule's boundary. For this purpose, a physics inspired weight map that takes the user input into account is proposed, which is used both as a feature map and in the system's loss function. Our approach is extensively evaluated on the public LIDC-IDRI dataset, where we achieve a state-of-the-art performance of 0.55 intersection over union vs the 0.59 inter-observer agreement. Also, we show that iW-Net allows to correct the segmentation of small nodules, essential for proper patient referral decision, as well as improve the segmentation of the challenging non-solid nodules and thus may be an important tool for increasing the early diagnosis of lung cancer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge