Alex Gaudio
ExplainFix: Explainable Spatially Fixed Deep Networks
Mar 18, 2023Abstract:Is there an initialization for deep networks that requires no learning? ExplainFix adopts two design principles: the "fixed filters" principle that all spatial filter weights of convolutional neural networks can be fixed at initialization and never learned, and the "nimbleness" principle that only few network parameters suffice. We contribute (a) visual model-based explanations, (b) speed and accuracy gains, and (c) novel tools for deep convolutional neural networks. ExplainFix gives key insights that spatially fixed networks should have a steered initialization, that spatial convolution layers tend to prioritize low frequencies, and that most network parameters are not necessary in spatially fixed models. ExplainFix models have up to 100x fewer spatial filter kernels than fully learned models and matching or improved accuracy. Our extensive empirical analysis confirms that ExplainFix guarantees nimbler models (train up to 17\% faster with channel pruning), matching or improved predictive performance (spanning 13 distinct baseline models, four architectures and two medical image datasets), improved robustness to larger learning rate, and robustness to varying model size. We are first to demonstrate that all spatial filters in state-of-the-art convolutional deep networks can be fixed at initialization, not learned.
HeartSpot: Privatized and Explainable Data Compression for Cardiomegaly Detection
Oct 05, 2022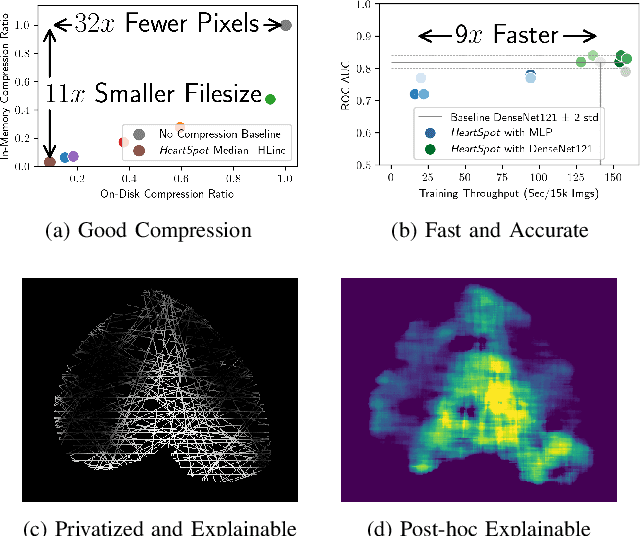
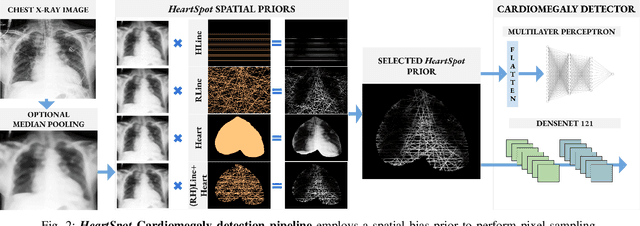

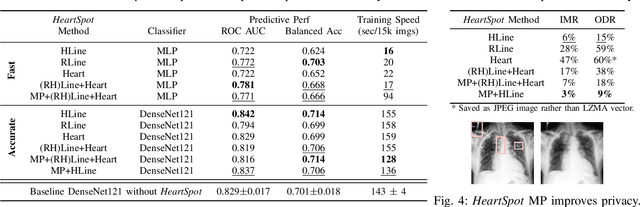
Abstract:Advances in data-driven deep learning for chest X-ray image analysis underscore the need for explainability, privacy, large datasets and significant computational resources. We frame privacy and explainability as a lossy single-image compression problem to reduce both computational and data requirements without training. For Cardiomegaly detection in chest X-ray images, we propose HeartSpot and four spatial bias priors. HeartSpot priors define how to sample pixels based on domain knowledge from medical literature and from machines. HeartSpot privatizes chest X-ray images by discarding up to 97% of pixels, such as those that reveal the shape of the thoracic cage, bones, small lesions and other sensitive features. HeartSpot priors are ante-hoc explainable and give a human-interpretable image of the preserved spatial features that clearly outlines the heart. HeartSpot offers strong compression, with up to 32x fewer pixels and 11x smaller filesize. Cardiomegaly detectors using HeartSpot are up to 9x faster to train or at least as accurate (up to +.01 AUC ROC) when compared to a baseline DenseNet121. HeartSpot is post-hoc explainable by re-using existing attribution methods without requiring access to the original non-privatized image. In summary, HeartSpot improves speed and accuracy, reduces image size, improves privacy and ensures explainability. Source code: https://www.github.com/adgaudio/HeartSpot
A Massively-Parallel 3D Simulator for Soft and Hybrid Robots
Jul 19, 2022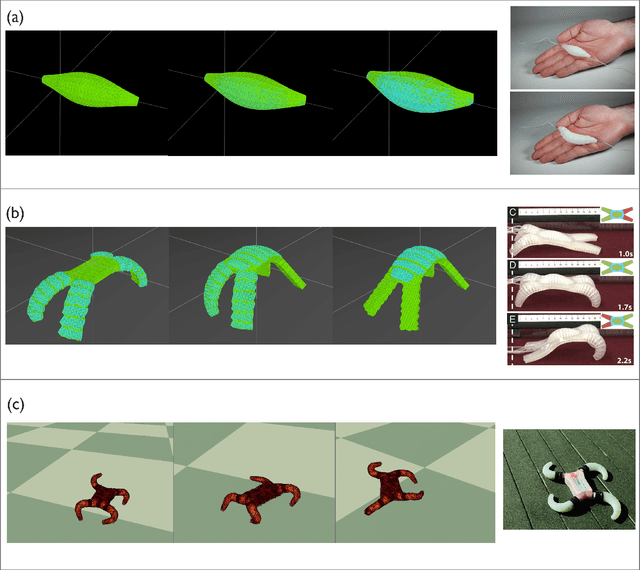

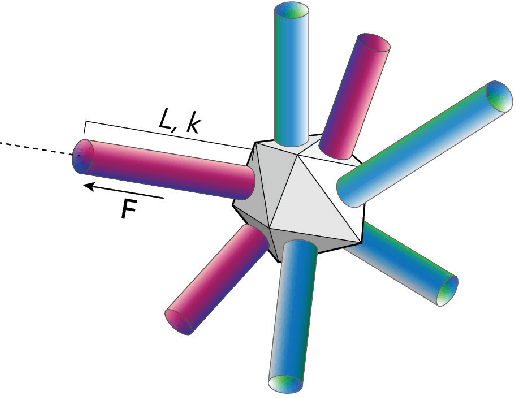
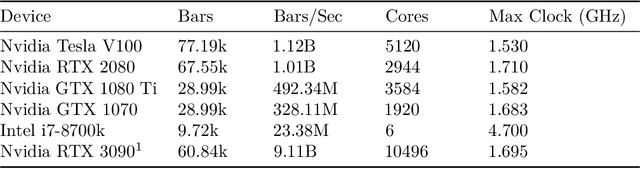
Abstract:Simulation is an important step in robotics for creating control policies and testing various physical parameters. Soft robotics is a field that presents unique physical challenges for simulating its subjects due to the nonlinearity of deformable material components along with other innovative, and often complex, physical properties. Because of the computational cost of simulating soft and heterogeneous objects with traditional techniques, rigid robotics simulators are not well suited to simulating soft robots. Thus, many engineers must build their own one-off simulators tailored to their system, or use existing simulators with reduced performance. In order to facilitate the development of this exciting technology, this work presents an interactive-speed, accurate, and versatile simulator for a variety of types of soft robots. Cronos, our open-source 3D simulation engine, parallelizes a mass-spring model for ultra-fast performance on both deformable and rigid objects. Our approach is applicable to a wide array of nonlinear material configurations, including high deformability, volumetric actuation, or heterogenous stiffness. This versatility provides the ability to mix materials and geometric components freely within a single robot simulation. By exploiting the flexibility and scalability of nonlinear Hookean mass-spring systems, this framework simulates soft and rigid objects via a highly parallel model for near real-time speed. We describe an efficient GPU CUDA implementation, which we demonstrate to achieve computation of over 1 billion elements per second on consumer-grade GPU cards. Dynamic physical accuracy of the system is validated by comparing results to Euler-Bernoulli beam theory, natural frequency predictions, and empirical data of a soft structure under large deformation.
Enhancement of Retinal Fundus Images via Pixel Color Amplification
Jul 28, 2020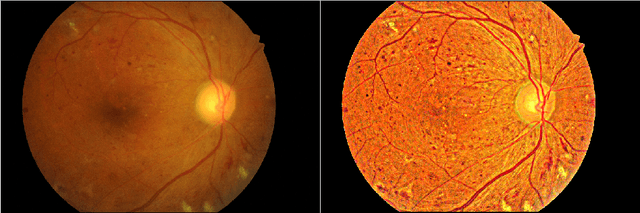

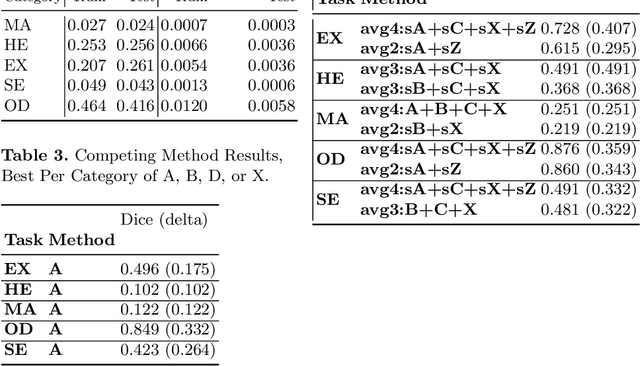
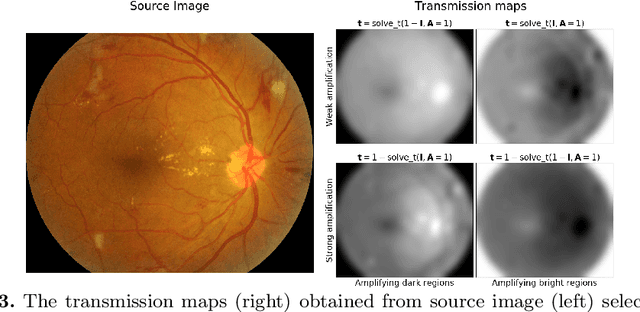
Abstract:We propose a pixel color amplification theory and family of enhancement methods to facilitate segmentation tasks on retinal images. Our novel re-interpretation of the image distortion model underlying dehazing theory shows how three existing priors commonly used by the dehazing community and a novel fourth prior are related. We utilize the theory to develop a family of enhancement methods for retinal images, including novel methods for whole image brightening and darkening. We show a novel derivation of the Unsharp Masking algorithm. We evaluate the enhancement methods as a pre-processing step to a challenging multi-task segmentation problem and show large increases in performance on all tasks, with Dice score increases over a no-enhancement baseline by as much as 0.491. We provide evidence that our enhancement preprocessing is useful for unbalanced and difficult data. We show that the enhancements can perform class balancing by composing them together.
Improving Lesion Segmentation for Diabetic Retinopathy using Adversarial Learning
Jul 27, 2020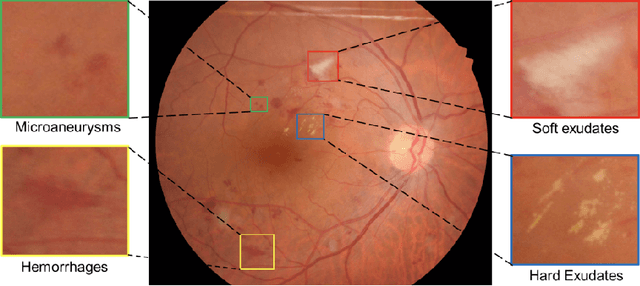
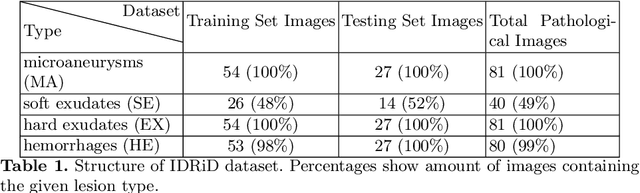
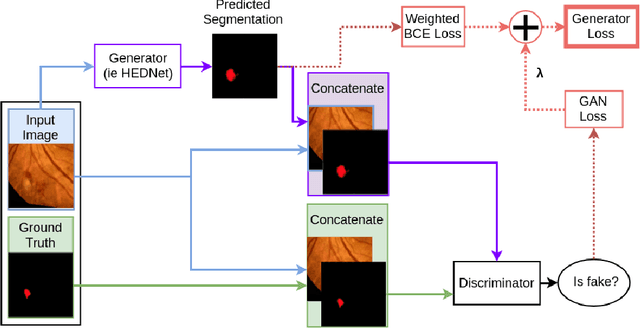
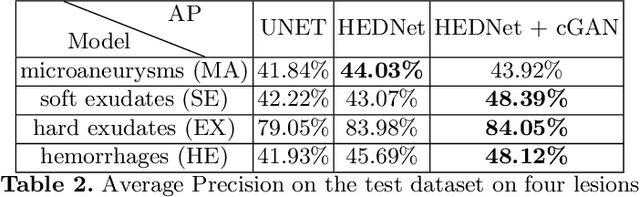
Abstract:Diabetic Retinopathy (DR) is a leading cause of blindness in working age adults. DR lesions can be challenging to identify in fundus images, and automatic DR detection systems can offer strong clinical value. Of the publicly available labeled datasets for DR, the Indian Diabetic Retinopathy Image Dataset (IDRiD) presents retinal fundus images with pixel-level annotations of four distinct lesions: microaneurysms, hemorrhages, soft exudates and hard exudates. We utilize the HEDNet edge detector to solve a semantic segmentation task on this dataset, and then propose an end-to-end system for pixel-level segmentation of DR lesions by incorporating HEDNet into a Conditional Generative Adversarial Network (cGAN). We design a loss function that adds adversarial loss to segmentation loss. Our experiments show that the addition of the adversarial loss improves the lesion segmentation performance over the baseline.
Learned Pre-Processing for Automatic Diabetic Retinopathy Detection on Eye Fundus Images
Jul 27, 2020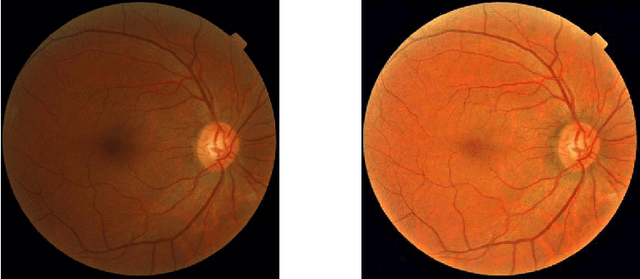
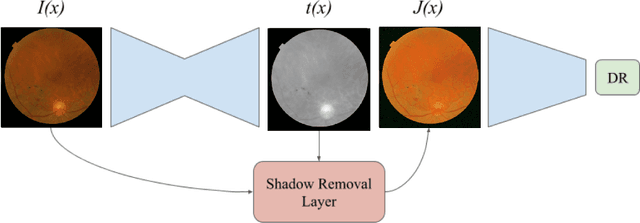
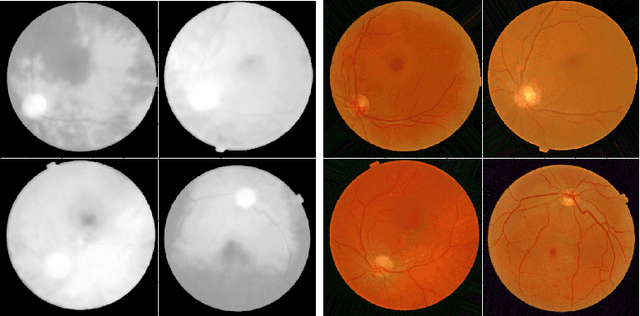
Abstract:Diabetic Retinopathy is the leading cause of blindness in the working-age population of the world. The main aim of this paper is to improve the accuracy of Diabetic Retinopathy detection by implementing a shadow removal and color correction step as a preprocessing stage from eye fundus images. For this, we rely on recent findings indicating that application of image dehazing on the inverted intensity domain amounts to illumination compensation. Inspired by this work, we propose a Shadow Removal Layer that allows us to learn the pre-processing function for a particular task. We show that learning the pre-processing function improves the performance of the network on the Diabetic Retinopathy detection task.
O-MedAL: Online Active Deep Learning for Medical Image Analysis
Aug 28, 2019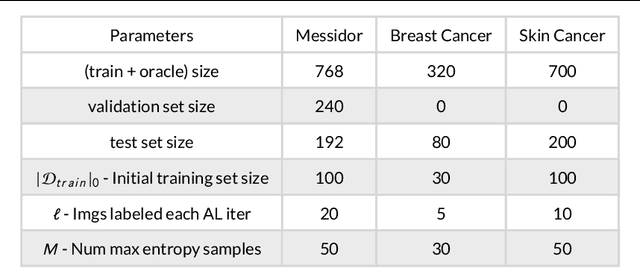
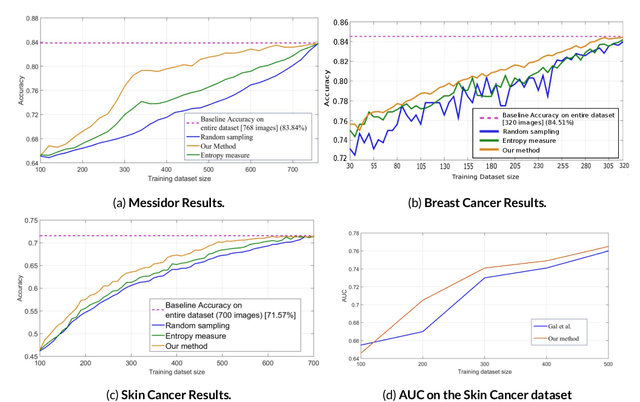
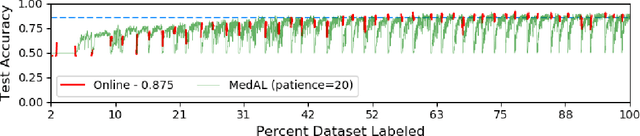
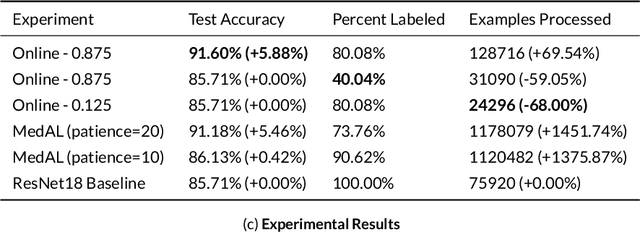
Abstract:Active Learning methods create an optimized and labeled training set from unlabeled data. We introduce a novel Online Active Deep Learning method for Medical Image Analysis. We extend our MedAL active learning framework to present new results in this paper. Experiments on three medical image datasets show that our novel online active learning model requires significantly less labelings, is more accurate, and is more robust to class imbalances than existing methods. Our method is also more accurate and computationally efficient than the baseline model. Compared to random sampling and uncertainty sampling, the method uses 275 and 200 (out of 768) fewer labeled examples, respectively. For Diabetic Retinopathy detection, our method attains a 5.88% accuracy improvement over the baseline model when 80% of the dataset is labeled, and the model reaches baseline accuracy when only 40% is labeled.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge