Alejo Nevado-Holgado
Bespoke Large Language Models for Digital Triage Assistance in Mental Health Care
Mar 28, 2024Abstract:Contemporary large language models (LLMs) may have utility for processing unstructured, narrative free-text clinical data contained in electronic health records (EHRs) -- a particularly important use-case for mental health where a majority of routinely-collected patient data lacks structured, machine-readable content. A significant problem for the the United Kingdom's National Health Service (NHS) are the long waiting lists for specialist mental healthcare. According to NHS data, in each month of 2023, there were between 370,000 and 470,000 individual new referrals into secondary mental healthcare services. Referrals must be triaged by clinicians, using clinical information contained in the patient's EHR to arrive at a decision about the most appropriate mental healthcare team to assess and potentially treat these patients. The ability to efficiently recommend a relevant team by ingesting potentially voluminous clinical notes could help services both reduce referral waiting times and with the right technology, improve the evidence available to justify triage decisions. We present and evaluate three different approaches for LLM-based, end-to-end ingestion of variable-length clinical EHR data to assist clinicians when triaging referrals. Our model is able to deliver triage recommendations consistent with existing clinical practices and it's architecture was implemented on a single GPU, making it practical for implementation in resource-limited NHS environments where private implementations of LLM technology will be necessary to ensure confidential clinical data is appropriately controlled and governed.
Developing Healthcare Language Model Embedding Spaces
Mar 28, 2024Abstract:Pre-trained Large Language Models (LLMs) often struggle on out-of-domain datasets like healthcare focused text. We explore specialized pre-training to adapt smaller LLMs to different healthcare datasets. Three methods are assessed: traditional masked language modeling, Deep Contrastive Learning for Unsupervised Textual Representations (DeCLUTR), and a novel pre-training objective utilizing metadata categories from the healthcare settings. These schemes are evaluated on downstream document classification tasks for each dataset, with additional analysis of the resultant embedding spaces. Contrastively trained models outperform other approaches on the classification tasks, delivering strong performance from limited labeled data and with fewer model parameter updates required. While metadata-based pre-training does not further improve classifications across the datasets, it yields interesting embedding cluster separability. All domain adapted LLMs outperform their publicly available general base LLM, validating the importance of domain-specialization. This research illustrates efficient approaches to instill healthcare competency in compact LLMs even under tight computational budgets, an essential capability for responsible and sustainable deployment in local healthcare settings. We provide pre-training guidelines for specialized healthcare LLMs, motivate continued inquiry into contrastive objectives, and demonstrates adaptation techniques to align small LLMs with privacy-sensitive medical tasks.
Efficiency at Scale: Investigating the Performance of Diminutive Language Models in Clinical Tasks
Feb 16, 2024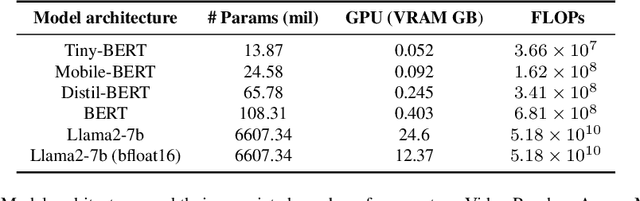
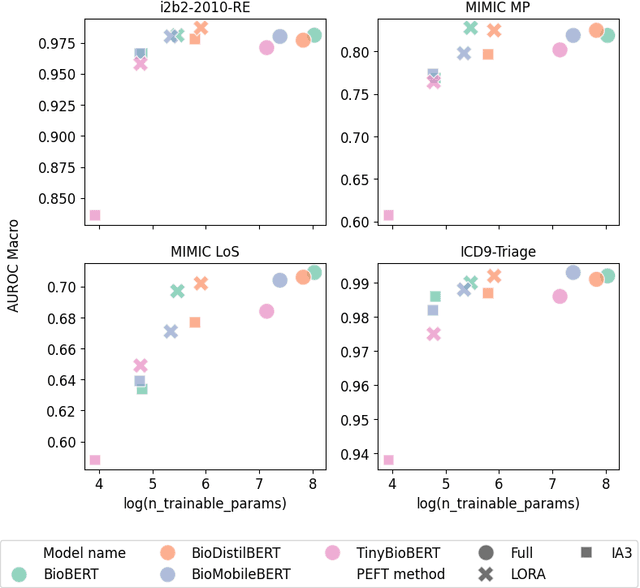
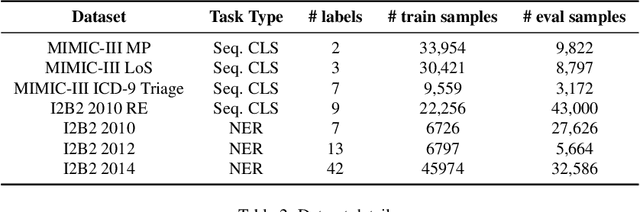
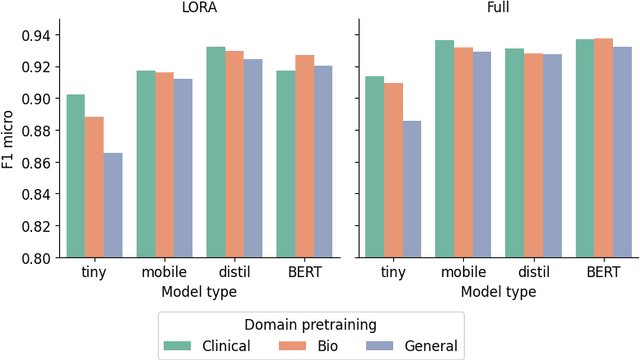
Abstract:The entry of large language models (LLMs) into research and commercial spaces has led to a trend of ever-larger models, with initial promises of generalisability, followed by a widespread desire to downsize and create specialised models without the need for complete fine-tuning, using Parameter Efficient Fine-tuning (PEFT) methods. We present an investigation into the suitability of different PEFT methods to clinical decision-making tasks, across a range of model sizes, including extremely small models with as few as $25$ million parameters. Our analysis shows that the performance of most PEFT approaches varies significantly from one task to another, with the exception of LoRA, which maintains relatively high performance across all model sizes and tasks, typically approaching or matching full fine-tuned performance. The effectiveness of PEFT methods in the clinical domain is evident, particularly for specialised models which can operate on low-cost, in-house computing infrastructure. The advantages of these models, in terms of speed and reduced training costs, dramatically outweighs any performance gain from large foundation LLMs. Furthermore, we highlight how domain-specific pre-training interacts with PEFT methods and model size, and discuss how these factors interplay to provide the best efficiency-performance trade-off. Full code available at: tbd.
Detecting the Clinical Features of Difficult-to-Treat Depression using Synthetic Data from Large Language Models
Feb 12, 2024



Abstract:Difficult-to-treat depression (DTD) has been proposed as a broader and more clinically comprehensive perspective on a person's depressive disorder where despite treatment, they continue to experience significant burden. We sought to develop a Large Language Model (LLM)-based tool capable of interrogating routinely-collected, narrative (free-text) electronic health record (EHR) data to locate published prognostic factors that capture the clinical syndrome of DTD. In this work, we use LLM-generated synthetic data (GPT3.5) and a Non-Maximum Suppression (NMS) algorithm to train a BERT-based span extraction model. The resulting model is then able to extract and label spans related to a variety of relevant positive and negative factors in real clinical data (i.e. spans of text that increase or decrease the likelihood of a patient matching the DTD syndrome). We show it is possible to obtain good overall performance (0.70 F1 across polarity) on real clinical data on a set of as many as 20 different factors, and high performance (0.85 F1 with 0.95 precision) on a subset of important DTD factors such as history of abuse, family history of affective disorder, illness severity and suicidality by training the model exclusively on synthetic data. Our results show promise for future healthcare applications especially in applications where traditionally, highly confidential medical data and human-expert annotation would normally be required.
Identifying DNA Sequence Motifs Using Deep Learning
Nov 20, 2023



Abstract:Splice sites play a crucial role in gene expression, and accurate prediction of these sites in DNA sequences is essential for diagnosing and treating genetic disorders. We address the challenge of splice site prediction by introducing DeepDeCode, an attention-based deep learning sequence model to capture the long-term dependencies in the nucleotides in DNA sequences. We further propose using visualization techniques for accurate identification of sequence motifs, which enhance the interpretability and trustworthiness of DeepDeCode. We compare DeepDeCode to other state-of-the-art methods for splice site prediction and demonstrate its accuracy, explainability and efficiency. Given the results of our methodology, we expect that it can used for healthcare applications to reason about genomic processes and be extended to discover new splice sites and genomic regulatory elements.
Personalised recommendations of sleep behaviour with neural networks using sleep diaries captured in Sleepio
Jul 29, 2022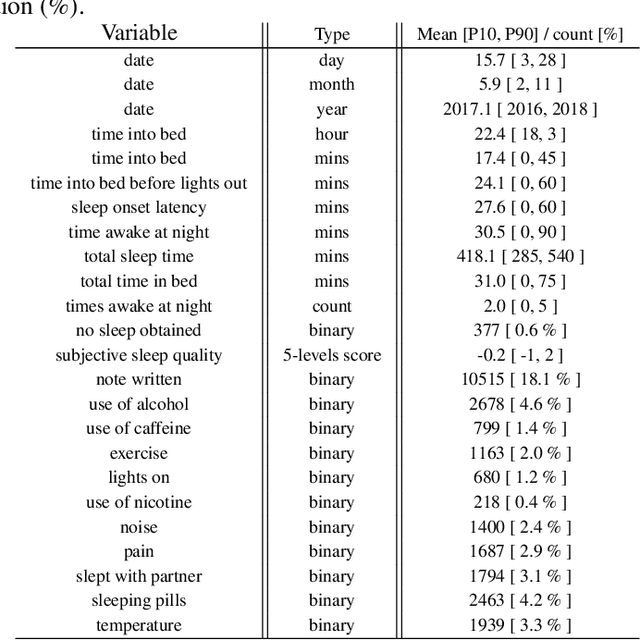
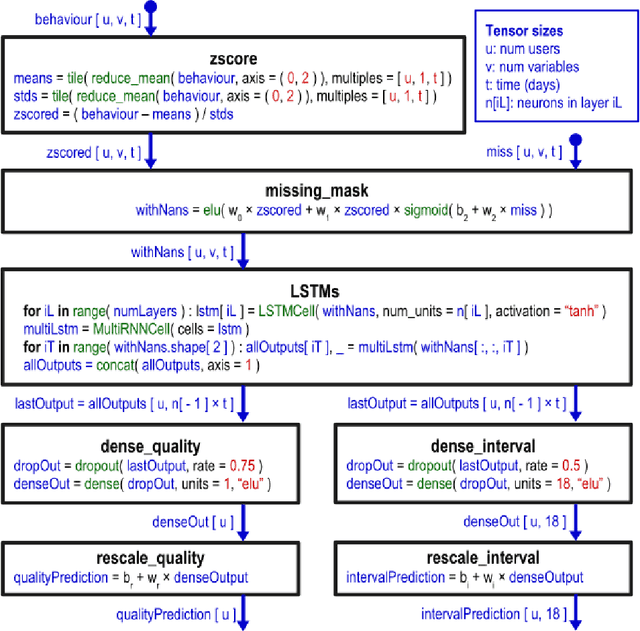
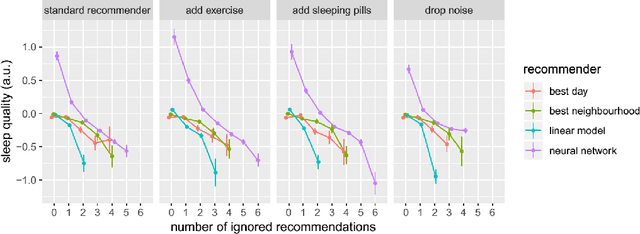
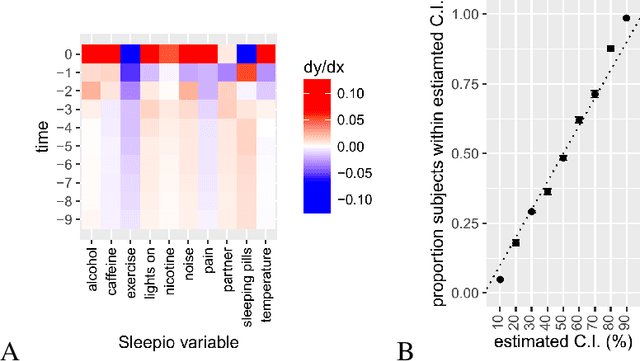
Abstract:SleepioTM is a digital mobile phone and web platform that uses techniques from cognitive behavioural therapy (CBT) to improve sleep in people with sleep difficulty. As part of this process, Sleepio captures data about the sleep behaviour of the users that have consented to such data being processed. For neural networks, the scale of the data is an opportunity to train meaningful models translatable to actual clinical practice. In collaboration with Big Health, the therapeutics company that created and utilizes Sleepio, we have analysed data from a random sample of 401,174 sleep diaries and built a neural network to model sleep behaviour and sleep quality of each individual in a personalised manner. We demonstrate that this neural network is more accurate than standard statistical methods in predicting the sleep quality of an individual based on his/her behaviour from the last 10 days. We compare model performance in a wide range of hyperparameter settings representing various scenarios. We further show that the neural network can be used to produce personalised recommendations of what sleep habits users should follow to maximise sleep quality, and show that these recommendations are substantially better than the ones generated by standard methods. We finally show that the neural network can explain the recommendation given to each participant and calculate confidence intervals for each prediction, all of which are essential for clinicians to be able to adopt such a tool in clinical practice.
Clinical Prompt Learning with Frozen Language Models
May 11, 2022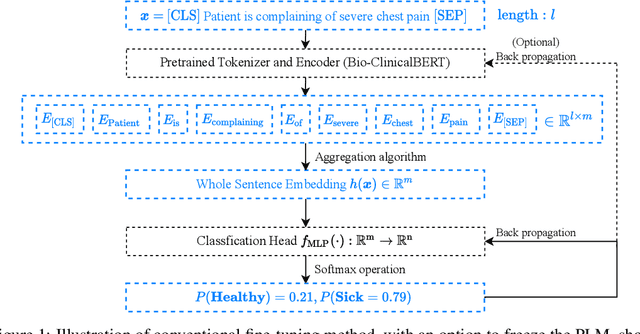
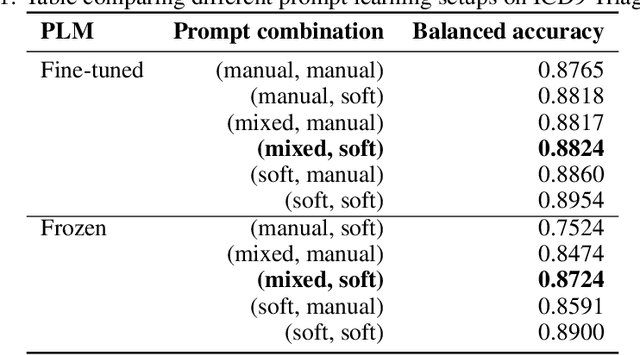
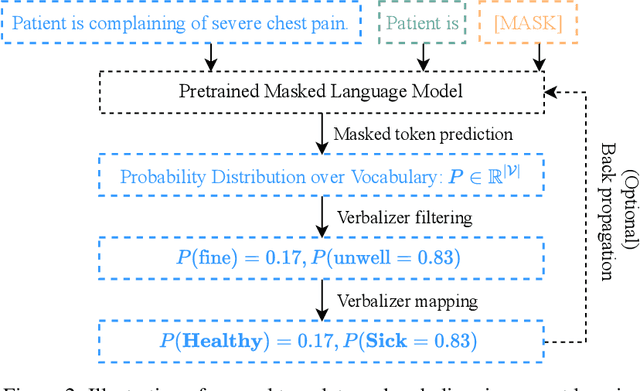
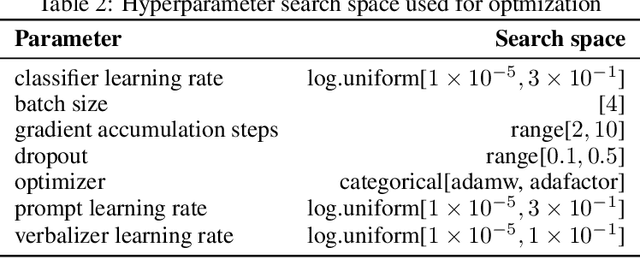
Abstract:Prompt learning is a new paradigm in the Natural Language Processing (NLP) field which has shown impressive performance on a number of natural language tasks with common benchmarking text datasets in full, few-shot, and zero-shot train-evaluation setups. Recently, it has even been observed that large but frozen pre-trained language models (PLMs) with prompt learning outperform smaller but fine-tuned models. However, as with many recent NLP trends, the performance of even the largest PLMs such as GPT-3 do not perform well on specialized domains (e.g. medical text), and the common practice to achieve State of the Art (SoTA) results still consists of pre-training and fine-tuning the PLMs on downstream tasks. The reliance on fine-tuning large PLMs is problematic in clinical settings where data is often held in non-GPU environments, and more resource efficient methods of training specialized domain models is crucial. We investigated the viability of prompt learning on clinically meaningful decision tasks and directly compared with more traditional fine-tuning methods. Results are partially in line with the prompt learning literature, with prompt learning able to match or improve on traditional fine-tuning with substantially fewer trainable parameters and requiring less training data. We argue that prompt learning therefore provides lower computational resource costs applicable to clinical settings, that can serve as an alternative to fine-tuning ever increasing in size PLMs. Complementary code to reproduce experiments presented in this work can be found at: https://github.com/NtaylorOX/Public_Clinical_Prompt.
Rationale production to support clinical decision-making
Nov 15, 2021



Abstract:The development of neural networks for clinical artificial intelligence (AI) is reliant on interpretability, transparency, and performance. The need to delve into the black-box neural network and derive interpretable explanations of model output is paramount. A task of high clinical importance is predicting the likelihood of a patient being readmitted to hospital in the near future to enable efficient triage. With the increasing adoption of electronic health records (EHRs), there is great interest in applications of natural language processing (NLP) to clinical free-text contained within EHRs. In this work, we apply InfoCal, the current state-of-the-art model that produces extractive rationales for its predictions, to the task of predicting hospital readmission using hospital discharge notes. We compare extractive rationales produced by InfoCal to competitive transformer-based models pretrained on clinical text data and for which the attention mechanism can be used for interpretation. We find each presented model with selected interpretability or feature importance methods yield varying results, with clinical language domain expertise and pretraining critical to performance and subsequent interpretability.
An efficient representation of chronological events in medical texts
Oct 24, 2020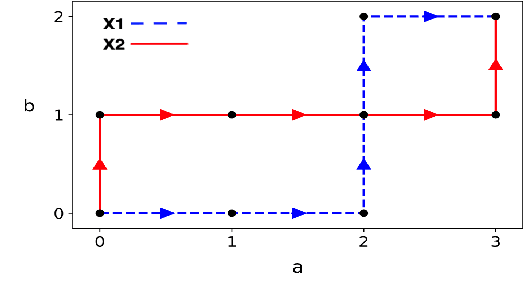
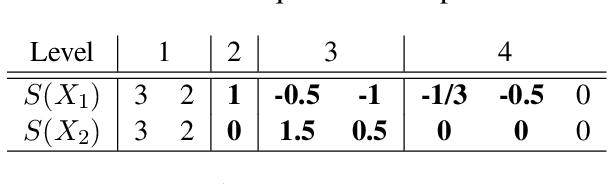

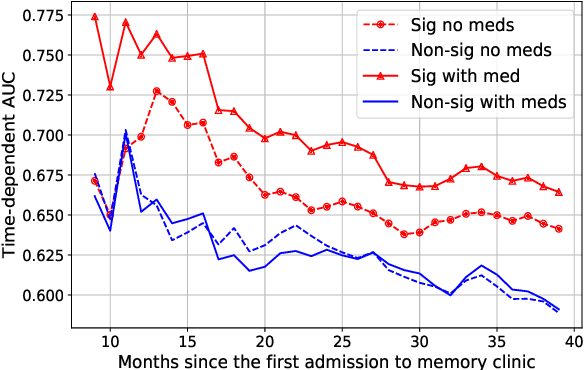
Abstract:In this work we addressed the problem of capturing sequential information contained in longitudinal electronic health records (EHRs). Clinical notes, which is a particular type of EHR data, are a rich source of information and practitioners often develop clever solutions how to maximise the sequential information contained in free-texts. We proposed a systematic methodology for learning from chronological events available in clinical notes. The proposed methodological {\it path signature} framework creates a non-parametric hierarchical representation of sequential events of any type and can be used as features for downstream statistical learning tasks. The methodology was developed and externally validated using the largest in the UK secondary care mental health EHR data on a specific task of predicting survival risk of patients diagnosed with Alzheimer's disease. The signature-based model was compared to a common survival random forest model. Our results showed a 15.4$\%$ increase of risk prediction AUC at the time point of 20 months after the first admission to a specialist memory clinic and the signature method outperformed the baseline mixed-effects model by 13.2 $\%$.
Population Gradients improve performance across data-sets and architectures in object classification
Oct 23, 2020



Abstract:The most successful methods such as ReLU transfer functions, batch normalization, Xavier initialization, dropout, learning rate decay, or dynamic optimizers, have become standards in the field due, particularly, to their ability to increase the performance of Neural Networks (NNs) significantly and in almost all situations. Here we present a new method to calculate the gradients while training NNs, and show that it significantly improves final performance across architectures, data-sets, hyper-parameter values, training length, and model sizes, including when it is being combined with other common performance-improving methods (such as the ones mentioned above). Besides being effective in the wide array situations that we have tested, the increase in performance (e.g. F1) it provides is as high or higher than this one of all the other widespread performance-improving methods that we have compared against. We call our method Population Gradients (PG), and it consists on using a population of NNs to calculate a non-local estimation of the gradient, which is closer to the theoretical exact gradient (i.e. this one obtainable only with an infinitely big data-set) of the error function than the empirical gradient (i.e. this one obtained with the real finite data-set).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge